Figures & data
Figure 1. Protein expression and isolation of the recombinant hMCM2-7 complex. (A) The complex was isolated by tandem affinity purification using a His-Tag and a Strep-Tag was loaded onto a Supedex200 HR10/30 column. The complex eluted as a single peak at 550 kDa. (B) 15 % SDS-PAGE of the complex showing the different subunits grouped in pairs. (C) Dynamic Light scattering of the hMCM2-7 complex shows the homogeneity of the heterohexamer. Polydispersity of hMCM2-7 shows a sample having a hydrodynamic radius of 100 Å in agreement with the heterohexamer size. The inserted table shows the statistics of the sample measured. (D) Western blot of the isolated hMCM2-7 complex using specific antibodies against the human MCM subunits. (E) Helicase assay demonstrates that the recombinant hMCM2-7 displays low activity.
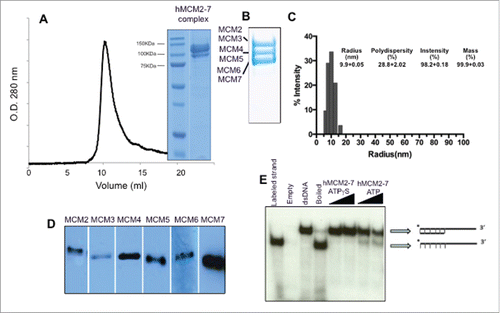
Figure 2. Electron microscopy fields of the hMCM2-7 in the absence of nucleotide or after incubation with ADP, ATPγS and ATPγS–DNA. The bar in each display represents 50 nm.
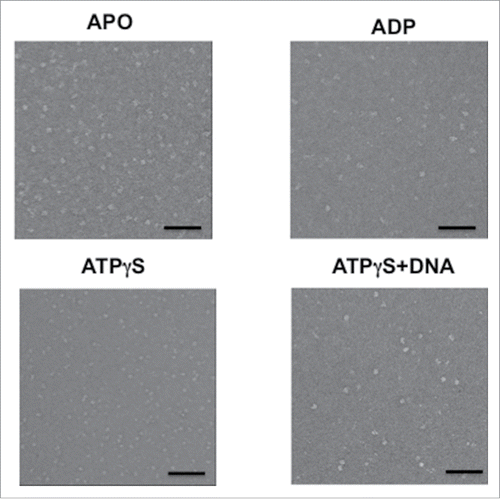
Figure 3. Detailed view of the negative stained preparations of the hMCM2-7 complex in the presence of different nucleotides. Gallery of raw particle images corresponding to top/bottom view (top row) of the apo (A), ADP (B), ATPγS (C) and ATPγS–DNA (D) preparations; reference free class-averages (middle raw); representative 2D class averages for each condition (lower raw). (E) Schematic representation of MCM2-7 complex stabilization as a consequence of binding of nucleotides and DNA.
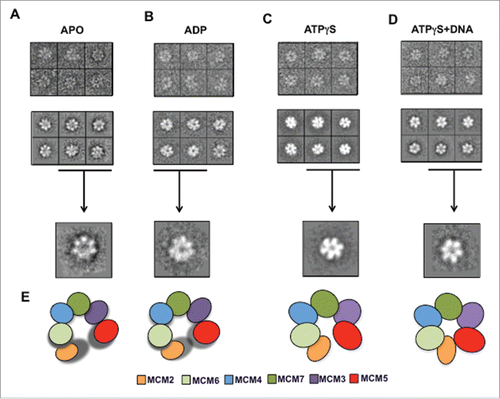
Figure 4. 3D reconstruction of hMCM2-7–ATPγS. (A) hMCM2-7–ATPγS reference-free class averages (left panel) and corresponding reprojections from the final structure (right panel). (B) Several views of different surface representations of a 3D reconstruction of hMCM2-7–ATPγS hexamer filtered to 28 Å. (C) Fourier Shell Correlation (FSC) curve of the 3D reconstructed map. The arrow indicates the MCM2-MCM5 “gate.”
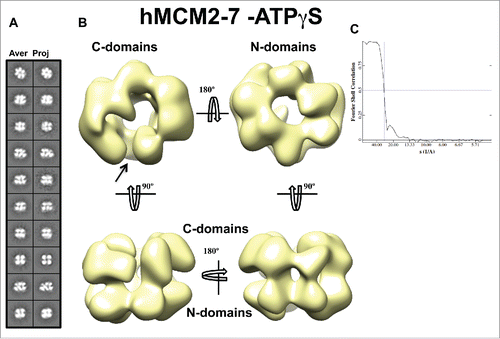
Figure 5. 3D reconstruction of the hMCM2-7–ATPγS–DNA complex. (A) hMCM2-7–ATPγS-DNA reference-free class averages (left panel) and corresponding reprojections from the final structure (right panel). (B) Several views of different surface representations of a 3D reconstruction of hMCM2-7–ATPγS-DNA hexamer filtered to 24 Å. (C) Fourier correlation shell graph of the 3D reconstructed map. The arrow indicates the MCM2-MCM5 “gate.”
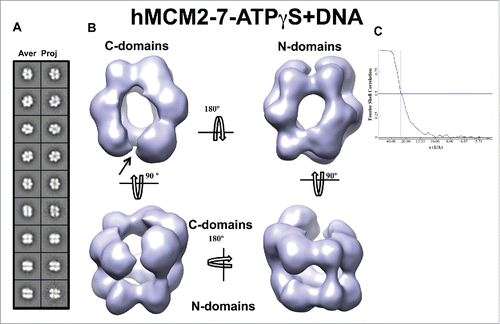
Figure 6. Comparison of the hMCM2-7 hexamer conformations. Top and side views of the volumes of hMCM2-7–ATPγS and hMCM2-7–ATPγS-DNA are compared to a 30 Å filtered map generated from the 3JA8 PDB file. The structure of the yeast MCM2-7 hexamer has been docked with Chimera to facilitate the comparison between the different maps and as a reference to assess the conformational changes of the different stages.
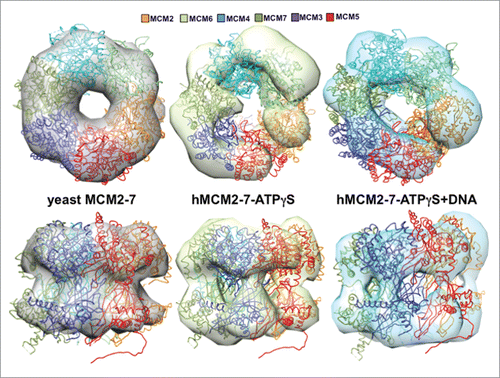
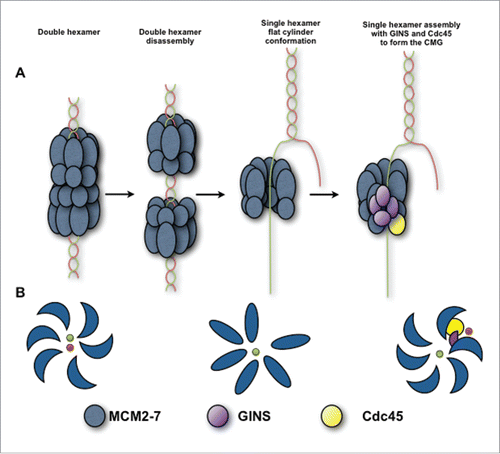
Figure 7. Schematic view of the hMCM2-7 conformations. (A) Eukaryotic MCM2-7 are loaded to dsDNA as double hexamer. Upon double hexamer disassembly and opening of dsDNA, MCM2-7 engages ssDNA. Subsequently GINS and Cdc45 would bind to MCM2-7 forming the active helicase complex, CMG. (B) Eukaryotic MCM2-7 in presence of nucleotide preferentially assumes a left-handed ring conformation. Upon ssDNA binding the MCM2-7 ring adopts more planar configuration. In the presence of GINS, Cdc45 and ssDNA, the AAA+ domain shifts again, but now in a right-handed spiral configuration. Hence, MCM2-7 experiences a chiral-flip in their conformation as it maturates into a functional helicase.