Figures & data
Figure 1. Inhibition of ORC/Cdc6/Cdt1 binding by MCM loaded on chromatin. (A) Plasmid DNA bound to magnetic beads was incubated in 10 µL of egg extract at 23˚C or at 4˚C for 16 min. For shifting the temperature from 4˚C to 23˚C, DNA beads were incubated at 4˚C for 16 min, and then shifted to 23˚C and incubated for the indicated time. DNA beads were isolated and washed with EB (50 mM HEPES-KOH at pH 7.5, 100 mM KCl, 2.5 mM MgCl2) containing 0.25% NP40 and bound proteins were analyzed by immunoblotting. (B) Scheme to examine the effect of MCM bound to DNA on the DNA binding of ORC/Cdc6/Cdt1 in the presence of geminin. (C) For the first incubation, DNA beads were incubated in 10 µL of egg extract in the presence or absence of geminin for the indicated time at 23˚C. DNA beads were then washed by either EB + 0.25% NP40 (low salt, L) or EB + 0.25% NP40 + 0.4 M NaCl (high salt, H) and transferred to the egg extracts (10 μL) containing geminin for the second incubation. After incubation at 23˚C for 20 min, the beads were washed with EB containing 0.25% NP40 (low salt) and bound proteins were analyzed by immunoblotting.
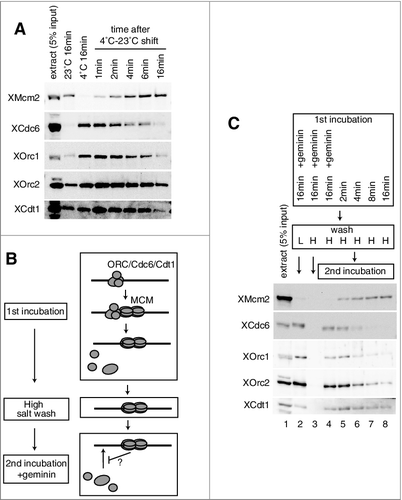
Figure 2. Inhibition of Cdc6, ORC, and Cdt1 binding by MCM on the plasmid. (A) The first incubation was performed, as in the legend for C, for 16 min in the presence or absence of geminin. After incubation, the beads were washed with the high-salt buffer, and further incubated in the second extract using either mock-, Mcm2-, Cdc6-, or Cdt1-depleted extracts in the presence of geminin. Immuno-depletion of each factor was performed by treating the egg extracts with protein A beads bound with either control, αXMcm2, αXCdt1, or αXCdc6 antisera. Various depleted extracts (0.5 uL) and proteins bound to the beads were analyzed by immunoblotting. (B) The first incubation was performed, as in the legend for C, for 16 min. The second incubation was performed in the extracts with or without 5 mM ATP-γ-S at either 23˚C or 4˚C in the presence of geminin.
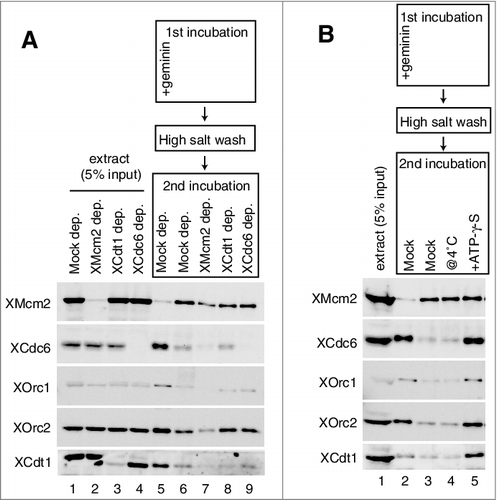
Figure 3. Inhibition of MCM and ORC/Cdc6/Cdt1 binding to DNA by the C-terminal fragment of MCM3. (A) Plasmid DNA bound to magnetic beads was incubated in 10 µL of egg extract at 23˚C in the absence (mock) and presence of 1.6 or 3.2 µM (∼ 5 × endogenous Mcm3) MCM3-C (His6-3Strep-XMcm3m(702-807)). After incubating for the indicated time, DNA beads were isolated and washed with EB + 0.25% NP40 and bound proteins were analyzed by immunoblotting. (B) DNA beads were incubated in 10 µL of egg extract for 16 min in the presence of various concentrations of MCM3-C as indicated. Beads were isolated and washed with either the low-salt or the high-salt buffer and bound Mcm2 was analyzed by immunoblotting. The graph shows quantified data of Mcm2 bound to DNA obtained by the low-salt wash (diamond) or the high-salt-wash (square). The vertical axis indicates the percentage of signal intensity relative to the maximum value obtained in the absence of MCM3-C. (C) DNA beads were incubated in 10 µL of egg extract in the presence or absence of geminin and MCM3-C (1.6 µM) for 20 min. Beads were isolated and washed with the low-salt EB buffer and the bound proteins were analyzed by immunoblotting. (D) DNA beads were incubated in 10 µL of egg extract in the presence of geminin for 15 min in the presence or absence of 1.6 µM MCM3-C (@0 min). Alternatively, DNA beads were incubated in the presence of geminin but the absence of MCM3-C for 15 min, and then MCM3-C (final concentration 1.6 μM) or the dialysis buffer was added and further incubated for 15 min (@15min). Beads were isolated and washed with the low-salt buffer and the bound proteins were analyzed by immunoblotting.
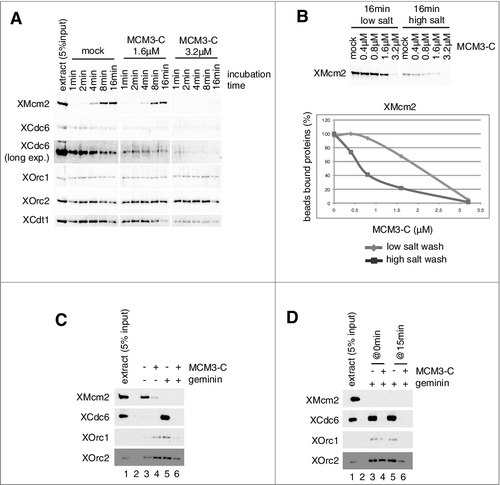
Figure 4. Inhibition of Cdc6 and ORC binding to DNA by MCM3-C under various conditions. (A and B) Experiments were performed with mock-, Mcm2-, Cdt1- and Cdc6 depleted extracts as described in the legend for C. (C) Experiments were performed as in the legend for C, except that DNA beads were incubated at either 23˚C or 4˚C. (D) Egg extracts were dialyzed with EB, and then supplemented with either 5 mM ATP or ATP-γ-S. The experiments that followed were performed as in the legend for C.
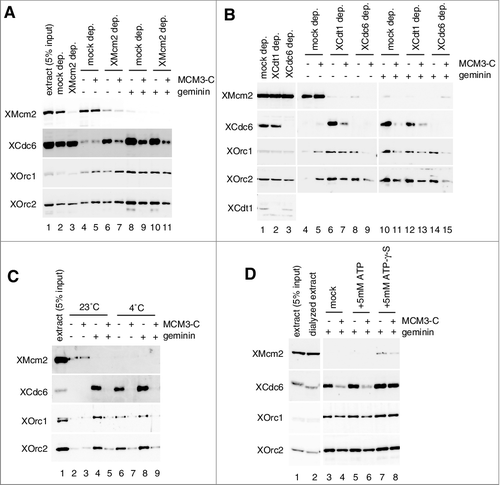
Figure 5. Functional region and amino acids of MCM3-C involved in the inhibition of Cdc6 binding to DNA. (A) Amino acid sequences of C-terminal regions of various metazoan Mcm3; Ciona intestinalis (Ci), Xenopus laevis maternal (Xlm), Danio rerio maternal (Drm), Xenopus laevis zygotic (Xlz), Danio rerio zygotic (Drz), Homo sapience (Hs), Gallus gallus (Gg). MCM3-C conserved motifs are further aligned with corresponding sequences of budding yeast (Sc) and C. elegans (Ce) Mcm3. (B) Experiments were performed as in the legend for C except that either maternal (XMCM3m-C aa 702–807 ( = MCM3-C)) or zygotic (XMCM3z-C aa 700–806) MCM3-C was used. (C and D) Experiments were performed as in the legend for C except that either 0.8 µM of various truncation mutants (C) or various point mutants (D) were used.
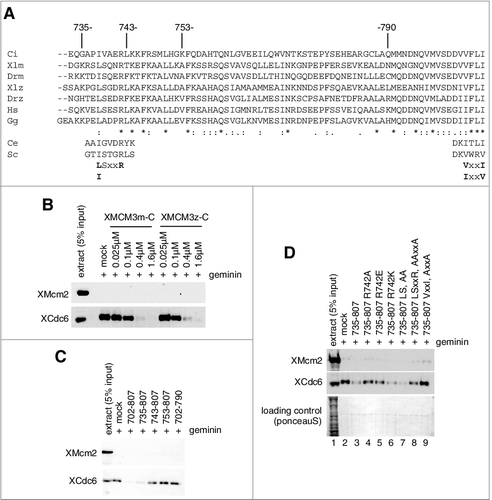
Figure 6. Requirement of soluble factor(s) in the extract for the dissociation of Cdc6/ORC by MCM3-C. (A) Experimental scheme for examining the requirement of soluble factor(s) for the dissociation of Cdc6 and ORC by MCM3-C. (B) DNA beads were incubated in 10 µL of egg extract containing geminin at 23˚C for 20 min (first incubation). Beads were then isolated and washed with the low-salt buffer and resuspended with EB or EB + 1mM ATP with or without 1.6 µM MCM3-C. After 20 min incubation at 23˚C (second incubation), supernatant and beads were separated and analyzed by immunoblotting following SDS-PAGE. (C) Experiments were performed as in (B) except that EB was supplemented with either egg extract or heat-treated supernatant (heat sup) at various ratios (%) for the second incubation. The heat sup was obtained by incubating egg extract at 98˚C for 2 min and centrifuging at 15,000 g for 15 min.
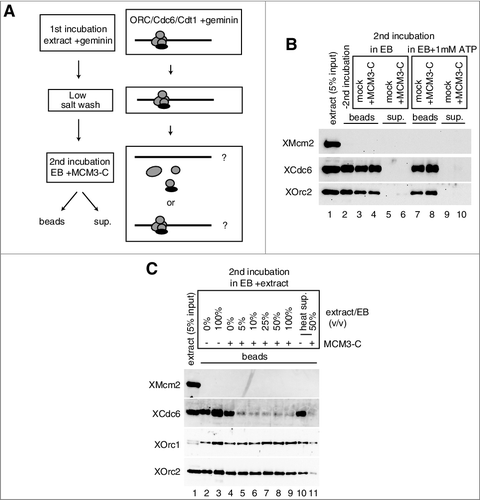
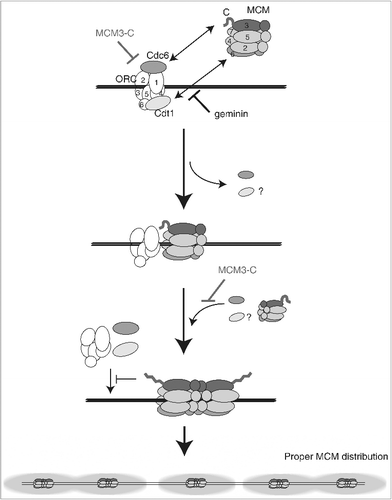
Figure 7. Model for the inhibitory autoregulation of MCM loading in the extract. Single hexameric MCM is recruited to DNA by prebound ORC/Cdc6/Cdt1 via interaction between MCM3 C-terminal-Cdc6 and between MCM6-Cdt1. Bound Cdc6 and Cdt1 are released on MCM loading, and the second MCM loading is promoted by rebinding of Cdc6 and Cdt1. After double hexamer formation, the conformation of a C-terminal region of MCM3 has changed and this inhibitory conformation could dissociate ORC/Cdc6/Cdt1 from DNA or inhibit their rebinding. The MCM3-C mimics the inhibitory conformation and could inhibit the first binding of ORC/Cdc6/Cdt1, or the second binding step of Cdc6/Cdt1. The inhibitory effect of MCM3-C is suppressed by ATP-γ-S and requires soluble factor(s) in the extract.