Figures & data
Figure 1. TAp73 is expressed in brain. (a) mRNA levels of TAp73 in the indicated regions of the brain isolated from wild-type mice. KO is used as negative control (RNA derived from the brain of p73 KO mice). (b) The expression of p73 in different regions of human brain was determined using R2.
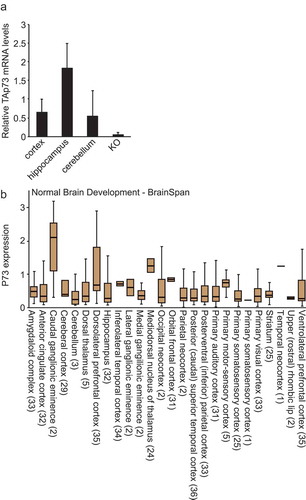
Figure 2. TAp73 KO mice maintain translation in the brain. (a) Brain slices obtained from the indicated genotypes were pulsed with puromycin (45 min; 5 μg/ml). Puromycin incorporation in the brain lysates was determined using Western blot and anti-puromycin antibodies as described [Citation25]. Scanned lanes were quantified using ImageJ. n = 3; *p < 0.05. (b) The levels of the indicated hippocampal proteins and their modifications in lysates obtained from the indicated genotypes were determined using Western blot. (c) Protein levels were quantified using ImageJ. n = 3; *p < 0.05. (d) The correlation between the expression of p73 and eEF2K in brain was determined using R2.
![Figure 2. TAp73 KO mice maintain translation in the brain. (a) Brain slices obtained from the indicated genotypes were pulsed with puromycin (45 min; 5 μg/ml). Puromycin incorporation in the brain lysates was determined using Western blot and anti-puromycin antibodies as described [Citation25]. Scanned lanes were quantified using ImageJ. n = 3; *p < 0.05. (b) The levels of the indicated hippocampal proteins and their modifications in lysates obtained from the indicated genotypes were determined using Western blot. (c) Protein levels were quantified using ImageJ. n = 3; *p < 0.05. (d) The correlation between the expression of p73 and eEF2K in brain was determined using R2.](/cms/asset/6cdb4375-36fe-4166-bc8b-32f327bc618b/kccy_a_1553341_f0002_c.jpg)
Figure 3. Proposed model for TAp73 interaction with the translation pathway. Under normal conditions, TAp73 promotes the translation of nucleolar proteins which process rRNA and thus promoting global protein synthesis. Acute TAp73 depletion results in reduced translation of nucleolar proteins, increased activity of eEF2K and reduced global protein synthesis. TAp73 KO in vivo triggers a compensation mechanism in the brain where eEF2K expression is reduced and global protein synthesis is maintained.