Figures & data
Figure 1. Overview of RNA-based control mechanisms employed by enteric pathogens of the family Enterobacteriaceae. mRNA translation can be controlled by RNA thermometers and by riboswitches within the 5′-UTR of target mRNAs in response to temperature or metabolites. Transcription, translation and/or stability of target transcripts can be modulated by cis-encoded asRNAs or trans-encoded ncRNAs. The RNA-binding protein CsrA modulates mRNA expression by interfering with translational initiation. The CsrB and CsrC RNAs counteract its activity. RNases control processing and degradation of ncRNAs and target transcripts. RNA-modifying enzymes change the efficiency of translation.
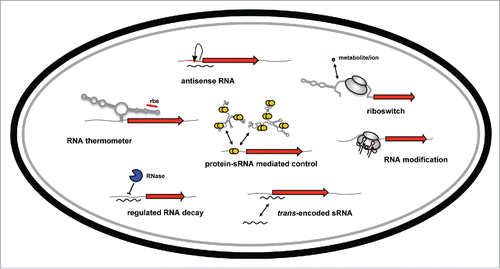
Table 1. Examples of virulence processes under RNA-based control.
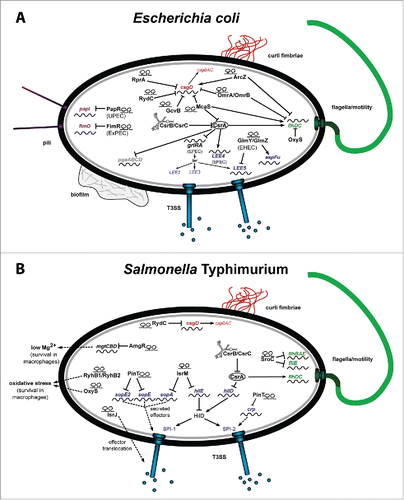
Figure 2. RNA-based control mechanism of E. coli and Salmonella. (A) Riboregulators controlling the colonization factors of pathogenic E. coli important for efficient cell adhesion and invasion. (B) Riboregulators coordinating the expression of Salmonella pathogenicity factors, responsible for cell entry, intracellular persistence and proliferation. The riboregulators are given in black and the target genes in green for flagella biosynthesis, in red for curli formation, in purple for pili production, and in blue for T3SS genes encoded on the pathogenicity islands LEE of EHEC/EPEC (A) or SPI-1 and SPI-2 of Salmonella (B).