Figures & data
Figure 1. GapA does not bind its own mRNA. DRaCALA with 60 fmol in vitro transcribed, [α-32P]UTP-labeled gapA operon mRNA was mixed with indicated amounts of GapA purified from a B. subtilis wild-type or Δsr1 strain as described in Materials and Methods.
![Figure 1. GapA does not bind its own mRNA. DRaCALA with 60 fmol in vitro transcribed, [α-32P]UTP-labeled gapA operon mRNA was mixed with indicated amounts of GapA purified from a B. subtilis wild-type or Δsr1 strain as described in Materials and Methods.](/cms/asset/351f7e09-7420-42c5-8e63-9bedc72cef53/krnb_a_1208894_f0001_b.gif)
Figure 2. Localization of gapA operon RNA regions dependent on stabilization by GapA/SR1P. Schematic representation of the analyzed gapA operon mutants. Mutants were integrated into the amyE locus of B. subtilis MG1P (Δsr1::phleo; ΔgapA::ery). For pMCG8 and pMCG13 that do not encode a functional gapA gene, strain MG2P (Δsr1::phleo; ΔgapA::ery, ΔthrC::gapA) was used. Strains were transformed with either inducible sr1 overexpression plasmid pWSR1 or empty vector pWH353. Cells were grown as described in Materials and Methods and SR1P dependent stabilization of mutated gapA operon RNA analyzed by Northern blotting (Fig. S1). Genes: boxed arrows (cggR: white; gapA: black; pgk’: gray); pcggR with 200 nt upstream region: hatched arrow; transcription start site: bent arrow; RNaseY processing site: vertical arrow; alternative gapA terminator: black hairpin; artificial bsrF terminator: gray hairpin. +, SR1P required for stabilization; -, RNAs stable in the absence of SR1P (concluded from 3 independent experiments).
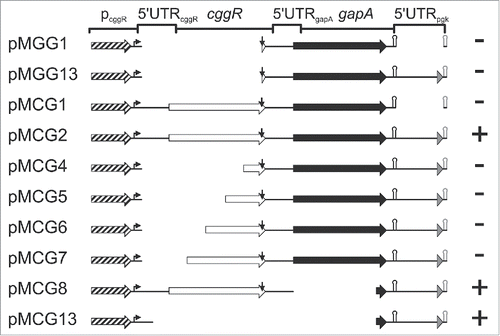
Figure 3. GapAStrep and GapA interact with RNases J1 and RNase Y. Far-Western blotting. A representative blot of 3 independently performed experiments is shown. Proteins were separated on 10% SDS-PAA gels and either stained with Coomassie (lanes 1–5) or blotted on PVDF membrane (lanes 6–17) as described in Materials and Methods. His-tagged RNases J1 and Y were purified from E. coli, GapAStrep was purified from B. subtilis DB104 (ΔamyE::gapAStrep) and untagged GapA was co-purified with Strep-tagged SR1P from B. subtilis Δsr1::cat (pWSR1/M25). (A) After blocking all blots were incubated with PBST gelatine (control, lanes 6–9) or PBST-gelatine containing either 170 µg RNase J1His (lanes 10–13) or 170 µg RNase YHis (lanes 14–17 RNase binding was detected with mouse anti-His-tag antibodies. Both RNases were able to bind GapAStrep and GapA. (B) Far Western Blot as in A) except that blots were incubated with 100 µg GapAStrep purified from either B. subtilis DB104 (ΔamyE::gapAStrep) (lanes 10–13) or B. subtilis DB104 (Δsr1::phleo, ΔamyE::gapAStrep) (lanes 14–17). GapAStrep binding was detected with mouse anti-Strep-tag antibodies.
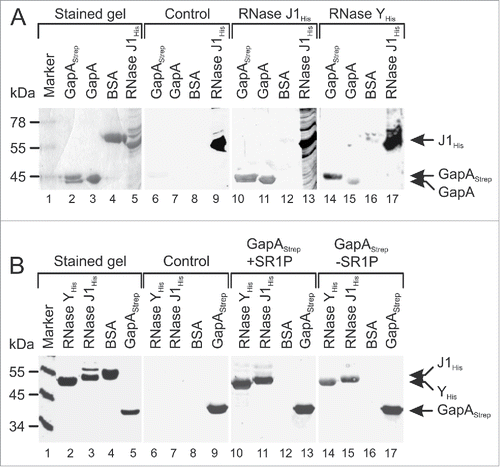
Figure 4. RNases J1 and Y co-purify with GapA. Co-elution assays Citation22 followed by Western blotting as described in Materials and Methods. A) Top: Western blot (WB) for detection of co-eluted RNase J1 within GapA preparations. GapAStrep was either purified from B. subtilis DB104 (ΔamyE::gapAStrep), DB104 (Δsr1::phleo; ΔamyE::gapAStrep) or DB104 (ΔrnjA::spec; ΔamyE::gapAStrep). To exclude that RNA bridges a possible interaction, an aliquot of the protein crude extracts was incubated with RNase A prior to protein purification (indicated by black asterisk). BSA and RNase J1His purified from E. coli serve as controls. RNase J specific antibodies were used. Bottom: Coomassie stained gel (CG) of the Western blot (loading control). The amount of protein loaded onto the gels is indicated. (B) Top: Western Blot (WB) as in (A) for detection of co-eluted RNase Y within GapA preparations. RNase Y specific antibodies were used. Bottom: CG of Western blot as loading control. The amount of protein loaded onto the gels is indicated.
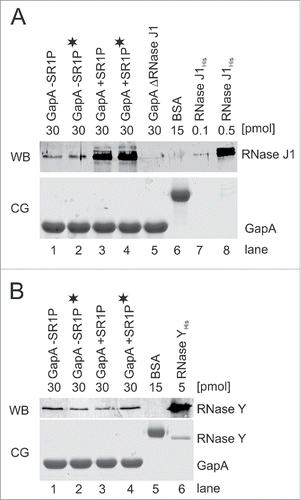
Figure 5. GapA/SR1P is involved in RNA degradation. RNA degradation assays with internally [α-32P]-UTP-labeled sRNA SR5. BSA and protein-free buffer were used as negative controls and 10 pmol RNase A as positive control. [γ-32P]-ATP-labeled pBR322xMspI served as size marker. Incubation was for 30 min at 37°C, followed by denaturation and separation on 8% denaturing polyacrylamide (PAA) gels, if not stated otherwise. (A) SR5 was incubated with RNase Y and RNase J1. The amount of protein used, full-length SR5 and degradation products (DP) are indicated. (B) Degradation assay as in A) with GapAStrep/SR1P purified from B. subtilis DB104 (ΔamyE::gapAStrep), GapAStrep purified from B. subtilis DB104 (Δsr1::phleo, ΔamyE::gapAStrep) and GapAStrep/SR1P purified from the ΔrnjA strain. (C) Time-course RNA degradation assay. SR5 was incubated with 10 pmol GapAStrep purified from Bacillus subtilis DB104 (ΔamyE::gapAStrep) or 10 pmol RNase J1His purified from E. coli. Incubation times and degradation products (DP) are indicated. (D) Degradation of the known RNase J1 target threonyl tRNA with purified RNases J1 and Y and GapAStrep +/−-SR1P. A schematic representation of the RNase J1 cleavage pattern of threonyl tRNA based on Citation33 is shown in Fig. S3. (E) Assay as in A) with GapAStrep purified from B. subtilis cells harvested from log- or stationary phase TY cultures. 10pmol of GapAStrep with or without co-purified SR1P were used. To exclude that the presence of SR1P increases an intrinsic RNase activity of GapA rather than that of co-purified RNase J1, SR1PHis was purified separately and added to the degradation assays. Neither SR1PHis alone (lane 8) nor addition of SR1PHis to SR1P-free GapAStrep (lanes 2 and 5) increased the RNase activity. SR1P, peptide co-purified with GapAStrep; SR1PHis; 50 pmol peptide purified separately and added later to purified GapAStrep.
![Figure 5. GapA/SR1P is involved in RNA degradation. RNA degradation assays with internally [α-32P]-UTP-labeled sRNA SR5. BSA and protein-free buffer were used as negative controls and 10 pmol RNase A as positive control. [γ-32P]-ATP-labeled pBR322xMspI served as size marker. Incubation was for 30 min at 37°C, followed by denaturation and separation on 8% denaturing polyacrylamide (PAA) gels, if not stated otherwise. (A) SR5 was incubated with RNase Y and RNase J1. The amount of protein used, full-length SR5 and degradation products (DP) are indicated. (B) Degradation assay as in A) with GapAStrep/SR1P purified from B. subtilis DB104 (ΔamyE::gapAStrep), GapAStrep purified from B. subtilis DB104 (Δsr1::phleo, ΔamyE::gapAStrep) and GapAStrep/SR1P purified from the ΔrnjA strain. (C) Time-course RNA degradation assay. SR5 was incubated with 10 pmol GapAStrep purified from Bacillus subtilis DB104 (ΔamyE::gapAStrep) or 10 pmol RNase J1His purified from E. coli. Incubation times and degradation products (DP) are indicated. (D) Degradation of the known RNase J1 target threonyl tRNA with purified RNases J1 and Y and GapAStrep +/−-SR1P. A schematic representation of the RNase J1 cleavage pattern of threonyl tRNA based on Citation33 is shown in Fig. S3. (E) Assay as in A) with GapAStrep purified from B. subtilis cells harvested from log- or stationary phase TY cultures. 10pmol of GapAStrep with or without co-purified SR1P were used. To exclude that the presence of SR1P increases an intrinsic RNase activity of GapA rather than that of co-purified RNase J1, SR1PHis was purified separately and added to the degradation assays. Neither SR1PHis alone (lane 8) nor addition of SR1PHis to SR1P-free GapAStrep (lanes 2 and 5) increased the RNase activity. SR1P, peptide co-purified with GapAStrep; SR1PHis; 50 pmol peptide purified separately and added later to purified GapAStrep.](/cms/asset/b4722e0c-e11c-4f2a-bdb5-402b18d2ec51/krnb_a_1208894_f0005_b.gif)
Figure 6. Determination of SR5 half-lives. B. subtilis strains 168, 168 (Δsr1::cat) and 168 (ΔgapA::ery) were grown in complex TY medium until onset of stationary growth phase, samples taken at the indicated times after rifampicin addition, total RNA prepared and separated on 6% denaturing PAA gels as described.Citation16 [α-32P]UTP-labeled riboprobes were used. Reprobing was performed with [γ-32P]ATP-labeled SB767 specific for 5S rRNA. Autoradiograms of Northern blots are shown. Half-lives presented under the gels are averaged from 3 independent determinations.
![Figure 6. Determination of SR5 half-lives. B. subtilis strains 168, 168 (Δsr1::cat) and 168 (ΔgapA::ery) were grown in complex TY medium until onset of stationary growth phase, samples taken at the indicated times after rifampicin addition, total RNA prepared and separated on 6% denaturing PAA gels as described.Citation16 [α-32P]UTP-labeled riboprobes were used. Reprobing was performed with [γ-32P]ATP-labeled SB767 specific for 5S rRNA. Autoradiograms of Northern blots are shown. Half-lives presented under the gels are averaged from 3 independent determinations.](/cms/asset/32bd5554-2e07-466e-8e5f-3ce0a6645fc0/krnb_a_1208894_f0006_b.gif)
Figure 7. Determination of gapA mRNA half-life. B. subtilis strains 168, 168 (ΔrnjA::spec); 168 (ΔrnjB::ery) were grown in complex TY medium until onset of stationary growth phase, samples taken at the indicated times after rifampicin addition, total RNA prepared and separated on 1.5% agarose gels, probed and reprobed as described.Citation22 Autoradiograms of representative Northern blots are shown. Half-lives are averaged from at least 2 independent determinations.
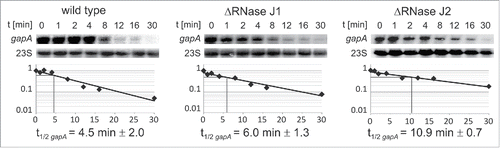
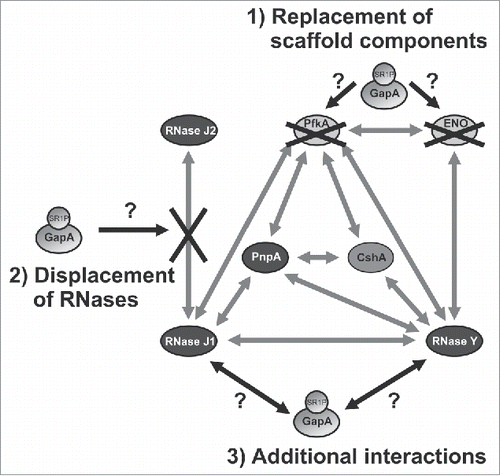
Figure 8. Working model on the potential function(s) of GapA/SR1P in the B. subtilis degradosome. Illustration of the possible effects of GapA/SR1P on the integrity of the B. subtilis degradosome. The interaction of GapA/SR1P might lead to 1) the replacement of the metabolic enzymes PfkA or enolase; 2) the displacement of RNase J2 from its interaction with RNase J1, or 3) additional interactions with RNase J1 and Y. In all cases the presence of GapA/SR1P within the degradosome might result in modulation of the RNase activity or specificity of the RNA degradation machinery. Gray arrows: interactions between the components of the RNA degradosome as observed by Commichau et al., 2009 Citation25 and Lehnik-Habrink et al., 2010,Citation48 black arrows and black crosses: possible effects of GapA/SR1P; gray ovals: proteins.