Figures & data
Figure 1. A model for regulation of the GAL cluster by GAL lncRNA R-loops and Dbp2. (A) Glucose Repression When glucose is available, Dbp2 is localized in the nucleusCitation74 and prevents the accumulation of R-loops at the GAL gene clusterCitation53 via the assembly of Dbp2-dependent lncRNA-protein complexes.Citation56 This allows the successful docking of the Cyc8/Tup1 co-repressor complex and subsequent repression. (B) Carbon Source Switch During a switch from glucose to galactose, Dbp2 is actively relocalized to the cytoplasm via export through the nuclear pore complex (NPC).Citation74 This may alter lncRNP assembly and cause R-loop formation and spreading across the GAL cluster. R-loop formation likely interferes with binding of transcriptional repressors and the Cyc8/Tup1 corepressor complex, causing derepression of the GAL genes. (C) Galactose Induction In the presence of galactose, the Gal4 transcriptional activator is released from the Gal80 inhibitor (not pictured),Citation96 enabling recruitment of co-activators and RNA P II.Citation97 This results in altered chromatin structure, as evidenced by looping of the GAL10 promoter and terminator,Citation53 which may enhance transcriptional induction. It is currently unknown how the GAL lncRNAs are cleared from chromatin or how Dbp2 is re-imported following long-term growth in galactose.
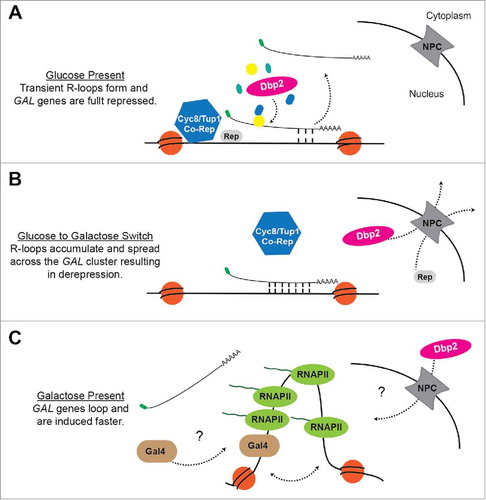
Table 1. Individually characterized lncRNAs of Saccharomyces cerevisiae. 65% of functionally characterized lncRNAs in S. cerevisiae have gene targets that function in pathways associated with metabolism or nutrient sensing/transport. AS = antisense. Us = upstream.
