Figures & data
Figure 1. Schematic overview of the organization and expression of immunoglobulin (Ig) genes. Different germline gene segments coding for the variable Ig heavy and light chains are joined by somatic V(D)J gene rearrangement (upper panels). Addition or removal of nucleotides during recombination at the junctions (symbolised by asterics) and somatic hypermutation (arrows) in the complementary-determining regions (CDR) of the VL and VH genes results in a high diversity of the Ig repertoire. The constant regions of the heavy chain are joined by RNA splicing to the variable regions. The heavy and light chains are covalently linked by disulfide bridges and fold into the typical Y-shaped immunoglobulin molecule. The antigen-binding site is formed by the CDRs of the heavy and light variable chains. A 3D shape of an Ig molecule can be found in the RCSB Protein Data Bank PDB ID: 1IGT (doi: 10.2210/pdb1igt/pdb).
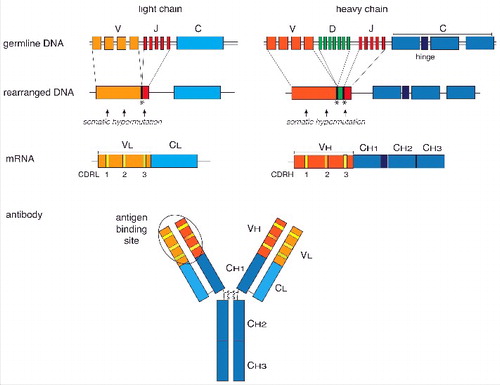
Figure 2. Conjugation of nucleosides to carrier proteins. In a first step the 2′ and 3′ hydroxyl groups of the ribose of the nucleic acid are oxidized with IO4− at pH9–9.5. This allows the coupling to primary amino groups of carrier proteins, e.g., ϵ-NH2-group of lysine residues. The resulting unstable acid is subsequently stabilized by reduction with NaBH4.
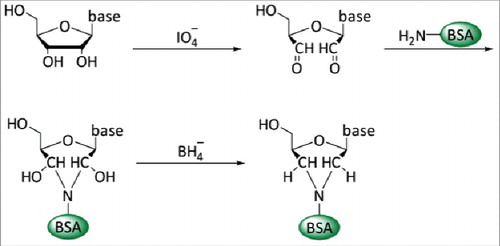
Table 1. Overview of the most commonly studied nucleic acid modifications and their analyses using antibody-based approaches.
