Figures & data
Figure 1. RNA exosome adaptors in S. cerevisiae and human nuclei. Principle overview of the main factors interacting with, and providing target specificity to, the exosome in S. cerevisiae (left) and human (right) nuclei (see text for details). hMTR4-Mtr4p (green) offers a main anchor linking the exosome to its different adaptors (indicated by black arrows). Question marks indicate possible interactions of Nsa1p and PICT1, UTP18 and DGCR8 with Mtr4p and hMTR4, respectively, motivated by protein homologies. Localization and function of C1D and MPP6 in the human nucleoplasm remain an open question (as question marks indicate). Note that proteins with the symbol ‘p’ derive from S. cerevisiae.
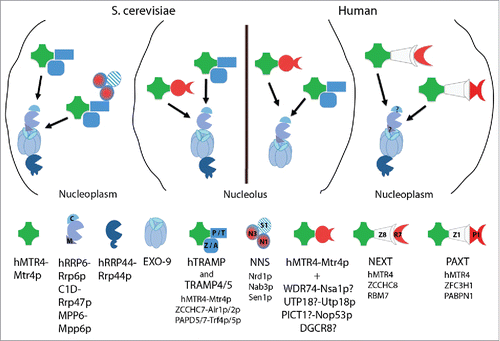
Figure 2. PABPs provide timing of RNA nuclear residence. Proceeding their poly(A) tail recognition by nuclear PABPs (red balloons), processed RNAs are exported to the cytoplasm (indicated by green light). In contrast, export-impaired or -delayed RNAs are turned over (red light). Thus, PABPs ‘sense’ the nuclear exposure time (represented by a the timer) of polyadenylated RNAs and provide the platform for nuclear RNA decay.
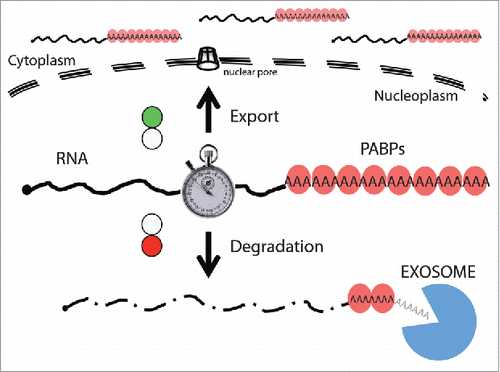
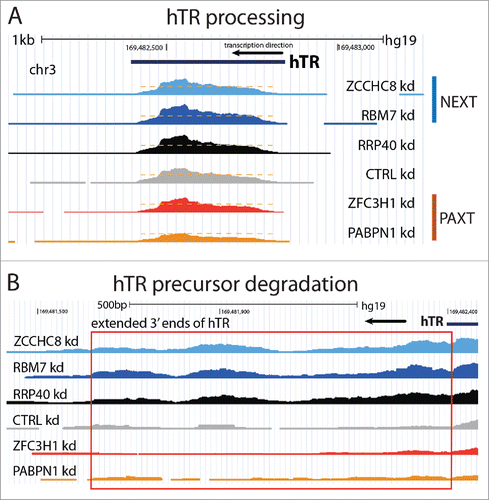
Figure 3. Processing and degradation of hTR. Screen shots of the hTR gene and RNAseq data from.Citation7 (A) hTR maturation depends on the PAXT-independent poly(A) tail recognition by PABPN1 and the ensuing 3′end trimming by PARN (see text for details). Accordingly, depletion of PABPN1 (‘PABPN1 Kd’), but not ZFC3H1 (‘ZFC3H1 kd’), decreases levels of mature hTR. Orange dashed lines indicate the maximum RNA signal levels in PABPN1 depletion conditions. (B) Extended hTR 3′ends (highlighted by red box) of impaired hTR precursors are targets for NEXT-dependent exosome degradation in the nucleoplasm.