Figures & data
Figure 1. Distinct biogenesis pathways mediate biogenesis of tRFs and tRNA halves. Active tRNA genes transcribed by the RNA Polymerase III complex produce a precursor tRNA transcript that is cleaved by RNAse P and Z to produce a mature tRNA transcript. This is further stabilized by the Lupus Autoantigen (La) that prevents entering of tRNAs into RNA silencing pathways. Entering of unstable tRNAs into RNAi pathways leads to the production of tRFs, although tRFs from the trailer regions of the pre-tRNA can also be formed independent of Dicer activity. Mature and stable tRNAs are exported to the cytoplasm where they are methylated by Dnmt2 to prevent cleavage by angiogenin. Angiogenin cleavage produces two fragments from mature tRNAs, derived from the 5’ or 3’ ends of the mature tRNA and termed 5’ or 3’ tRNA halves.
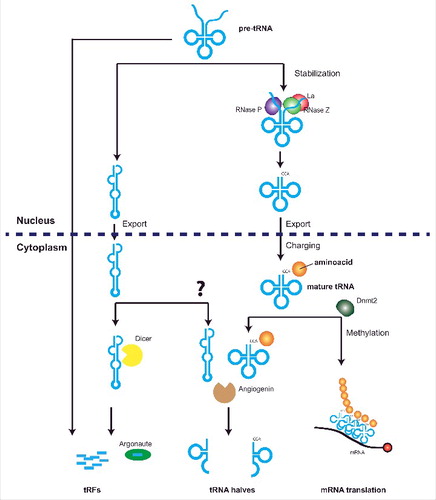
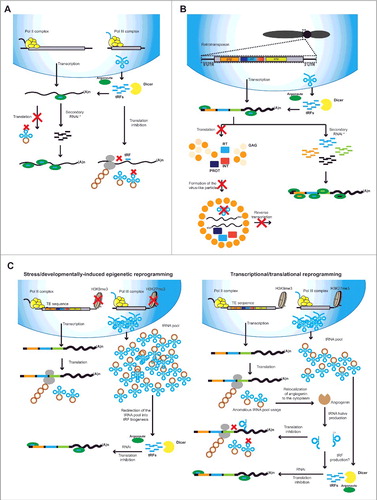
Figure 2. Known and speculative activity of tRFs. A. tRFs are known to act at two different levels: First, tRFs loaded into argonaute (AGO) can induce RNAi on targets mRNAs and potentially induce the initiation of secondary sRNA production. At the same time, degradation of tRNAs to tRFs inhibits the translation of mRNAs through a pathway yet to be identified. Translation inhibition could also take place due to degradation of mature tRNAs into tRFs and potential lack of mature tRNAs. B. Interaction of tRFs with the replication cycle of retrotransposons. tRFs loaded into AGO proteins are known to target retrotransposons and induce the production of secondary sRNAs from their RNA transcripts. At the same time, lack of mature tRNAs can inhibit translation of retrotransposon proteins and prevent reverse transcription. C. Proposed mechanisms linking TE reactivation and tRF biogenesis. Left panel: During strong epigenetic reprogramming, erasure of H3K9me3 and H3K27me3 leads to TE reactivation and excessive tRNA production. Excessive tRNA pool could be incorporated into RNAi and induce the production of tRFs. Right panel: Spurious reactivation of TE transcription can lead to an anomalous usage of the tRNA pool that if sensed by the cell can lead to angiogenin delocalization from the nucleus to the cytoplasm. This could lead to increased degradation of tRNAs into tRNA halves or direct tRF production. * Seconday RNAi will only take place in organisms with RdRPs like plants, fungi and invertebrate animals.