Figures & data
Figure 1. Canonical and non-canonical functions of eEF1A. During the elongation step of translation, ternary complexes of eEF1A, GTP and the different aminoacyl-tRNAs (aa-tRNA) are sampled by the ribosome. Upon productive base-pairing, the aa-tRNA is accommodated in the ribosome A-site while the eEF1A-bound GTP is hydrolyzed. eEF1A-GTP is regenerated through the guanosine exchange factor eEF1B. Non-canonical functions of eEF1A are indicated in the dashed box.
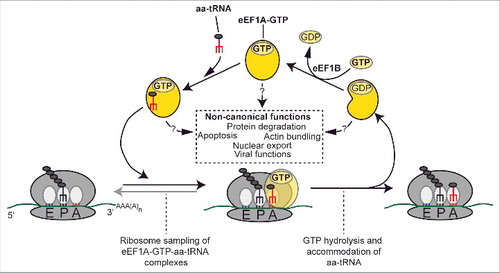
Figure 2. KMT-mediated lysine methylation. A lysine can accept up to three methyl groups through successive, KMT-mediated methylation. KMTs use S-adenosylmethionine (AdoMet), as methyl donor (not shown). Some KMT-mediated methylations can be reversed by lysine-specific demethylases (KDMs).
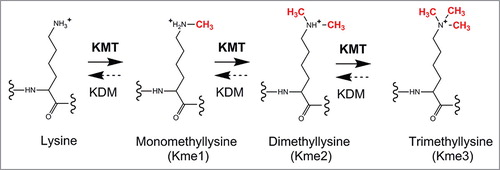
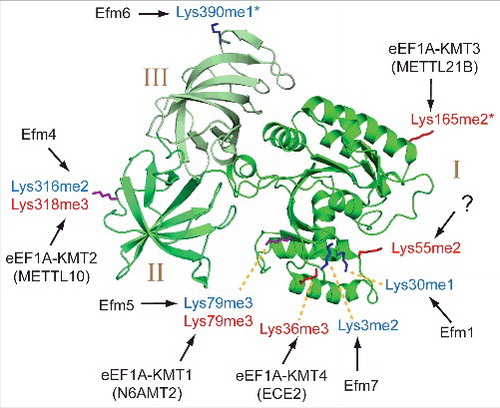
Figure 3. Methylated lysines and corresponding KMTs in yeast and human eEF1A. The three domains (I, II, III) of eEF1A are indicated in different shades of green colouring on a cartoon representation of the eEF1A structure. Methylated lysines in human and yeast eEF1A are indicated by red and blue labels, respectively, and also shown in stick representation, where methylation sites that are unique to human or yeast eEF1A are indicated in red and blue, respectively, whereas sites that are found in both proteins are shown in purple. The shown structure represents S. cerevisiae eEF1A (PDB 1F60), but since one of the methylated Lys residues in human eEF1A, Lys-165, is not conserved in yeast eEF1A, the corresponding residue (Ser-163) was, for the purpose of this illustration replaced by lysine (and indicated as human Lys-165). The KMTs responsible for lysine methylation in yeast and human (Efms and eEF1A-KMTs, respectively) are indicated. Asterisks indicate methylation sites that tend to display a mixture of different methylation states.