Figures & data
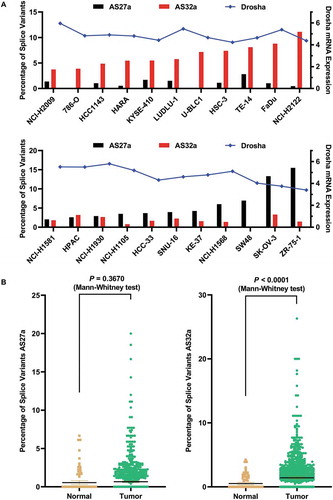
Figure 1. Identification of Drosha splice variants in human cells.
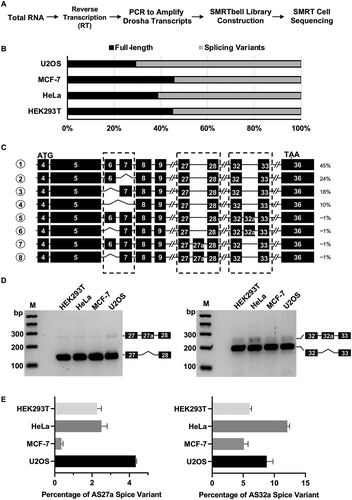
Figure 2. Characterization of Drosha-AS27a and Drosha-AS32a isoforms.
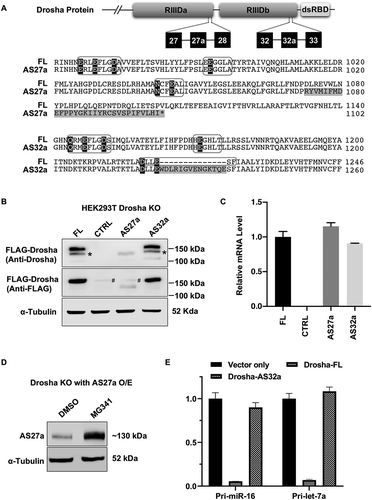
Figure 3. Drosha-AS27a and Drosha-AS32a are deficient in miRNA biogenesis.
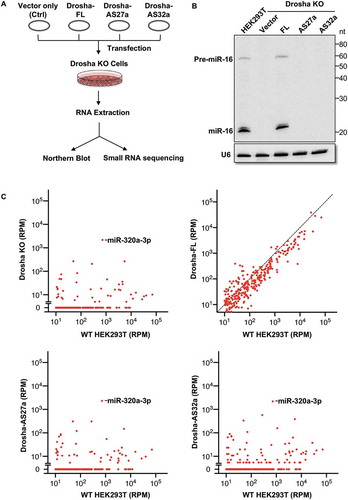
Figure 4. AS27a and AS32a are abundant in cancer cell lines and their levels are elevated in some breast cancer patients.