Figures & data
Figure 1. The dsRNA receptor SID-2 does not enhance dsRNA uptake for nutritional reasons
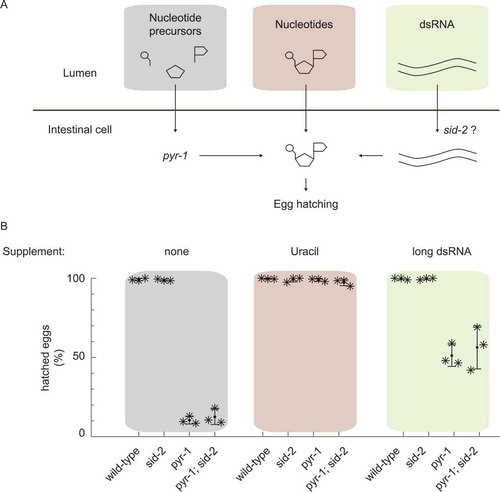
Figure 2. Sid-2 mutant animals grow faster and are elongated at hatching
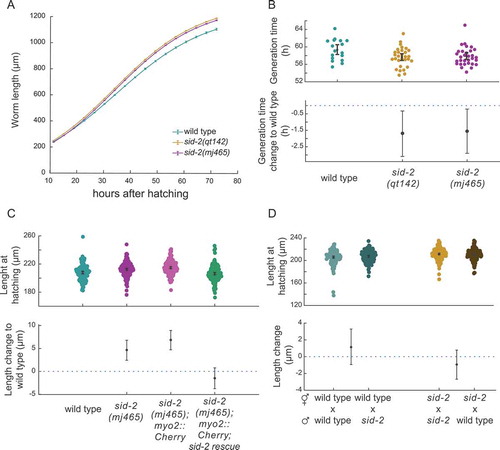
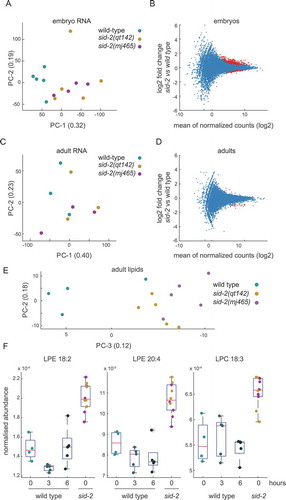
Figure 3. Sid-2 mutants show molecular phenotypes at embryonic and adult stage
Supplemental Material
Download Zip (19.2 MB)Availability of data
Sequencing data is available in the European Nucleotide Archive under the study accession number PRJEB32813.
