Figures & data
Table 1. Genome-wide profiling of discovery of miRNAs in molluscs
Figure 1. Genome-wide scanning of miRNAs in various species of molluscs (A). The relationship between the number of scanned miRNAs and genome size. (B). The genomic miRNA density in molluscs. The density of miRNA is defined as the ratio of the number of predicted miRNAs and the genome size (Mb). (C). Conserved miRNA family in molluscs. The numbers in the box indicate the isoforms of the predicted miRNAs in each family. (D). PCA of molluscs and other organisms based on the predicted miRNA family. Arth: arthropod Drosophila melanogaster (Dme); Fish: Teleostei fish Danio rerio (Dre); Mamm: mammal Mus musculus (Mmu) and Homo sapiens (Hsa); Nema: nematode Caenorhabditis elegans (Cel); Biva: Bivalvia; Gast: Gastropoda; Ceph: Cephalopoda. E. Venn diagram of miRNA candidates using small-RNA sequencing and genome-wide scanning in Crassostrea gigas F. Venn diagram of miRNA candidates using small-RNA sequencing and genome-wide scanning in Pinctada fucata.
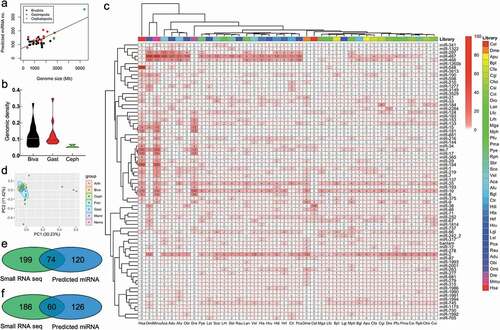
Figure 2. Conserved miR-184 in molluscs (A). Alignment of precursors of miR-184 in molluscs. miR-184 shows conserved mature sequences in molluscs. (B). Alignment of precursors of C. gigas miR-184 isoforms. (C). Structure of precursor of Cgi-miR-184.isoform.1. The underline shows mature miRNA
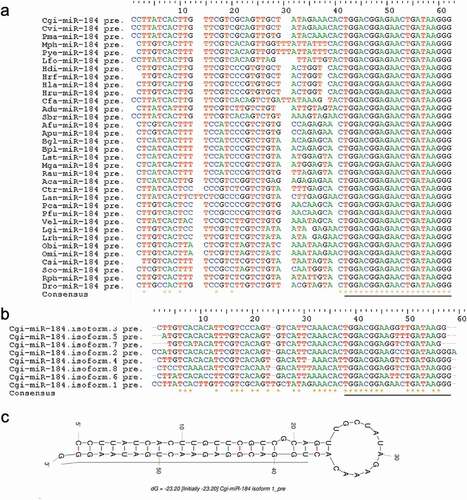
Figure 3. Genomic organization of miRNA genes in P. fucata (A). miRNA genes can be intergenic (alone or clustered) in the intron of non-coding RNA or protein coding genes (alone or clustered) or tandem duplicated on the genome. ‘+’ indicates miRNA genes transcribed from the sense strand, and ‘-’ indicates an antisense-strand assembly genome. (B). Composition of the various genomic organizations of miRNA genes in P. fucata. The majority of tandem duplicated miRNAs are located in intergenic region except for miR-29b located in intronic region
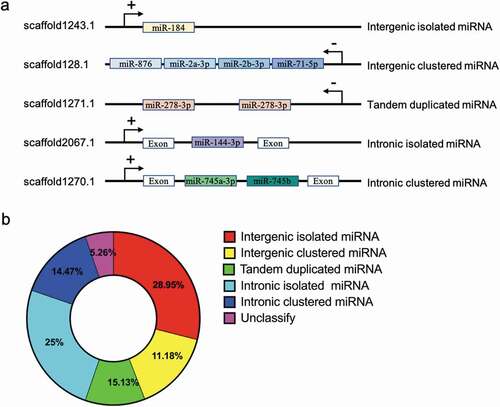
Figure 4. Target prediction of miRNAs in P. fucata (A). Venn diagram of the number of predicted target sites between genes and genomic identified miRNAs using miRanda and RNAhybrid algorithms. (B). The number of target genes for each miRNA. (C). miRNAs-target genes regulatory network of genomic-predicted miRNAs in P. fucata. (D). Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways, terms, and statistics of the KEGG pathway enrichment analysis for the predicted target genes of genomic miRNAs in P. fucata.
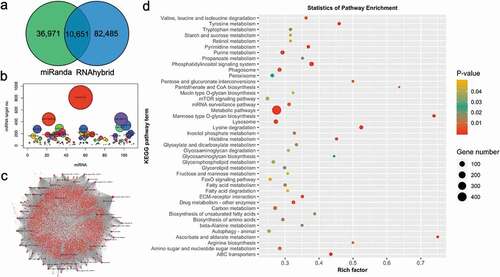
Table 2. miRNA biogenesis factors identified in molluscs based on genome scanning
Figure 5. miRNA biogenesis factors and pathway in molluscs (A). Conserved domains of miRNA biogenesis factors in P. fucata. (B). Phylogenetic relationships of key miRNA biogenesis factor AGO2 in molluscs. Inferred protein sequences were aligned using ClustalX, and conserved positions were extracted using Gblocks and subjected to phylogenetic analysis. Homologous sequences from vertebrates (Vert), including Danio rerio (Dre), Mus musculus (Mmu), and Homo sapiens (Hsa), were used as outgroups. Biva is an abbreviation for Bivalvia, which is shaded in green; Gast for Gastropoda in yellow; and Ceph for Cephalopoda in blue. (C). A graphical reconstruction of miRNA biogenesis in molluscs. The miRNAs are transcribed by RNA polymerase II (Pol II) into primary miRNAs (pri-miRNA) and then processed by the DROSHA-DGCR8 complex into precursor miRNAs (pre-miR). The mature miRNA strand is highlighted in red. The pre-miR was transported from the nucleus into the cytoplasm by XPO5-RAN complex, where it was then loaded into the DICER-TARBP2 complex and then into an miRNA duplex and unwound. The mature strand is incorporated into the RNA-induced silencing complex (miRISC). Depending on an miRNA’s complementarity to a target mRNA, the miRISC-mediated gene regulation will occur via either translational repression or mRNA degradation
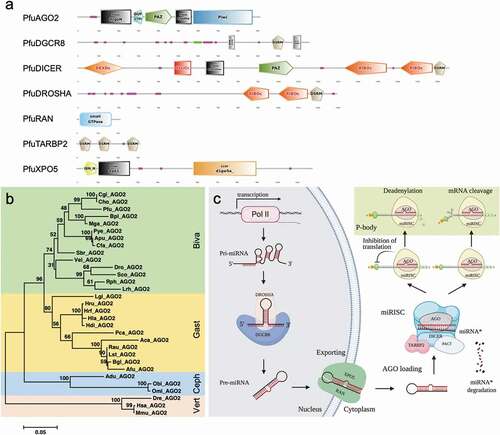
Table 3. Summary of identified miRNAs in molluscs
Table 4. Summary of validated miRNAs, their target genes, and functions in molluscs
