Figures & data
Table 1. List of primers used in the study
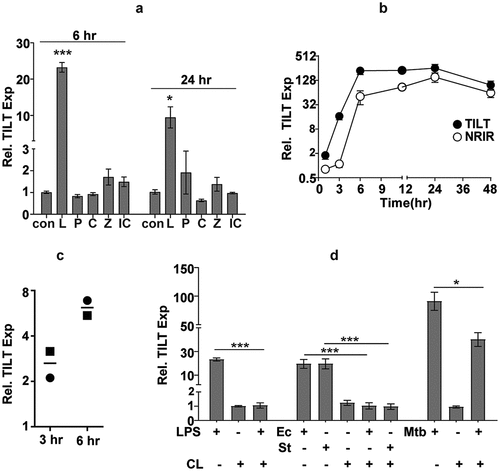
Figure 1. NRIR is expressed in response to bacterial infection
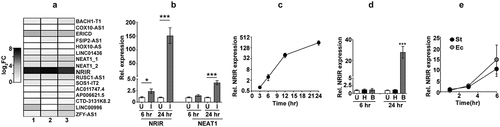
Figure 2. Identification of the novel transcripts-TILT encompassing CMPK2 and lncRNA – NRIR in Mtb infected macrophages
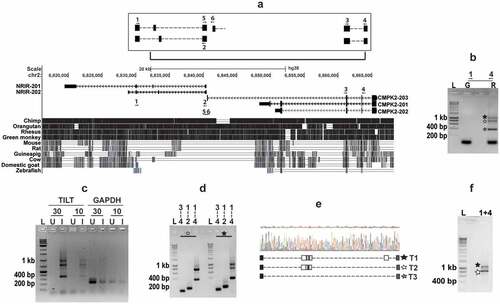
Figure 3. TILT is actively induced in Mtb infected macrophages
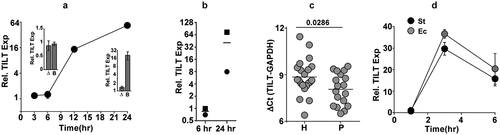
Figure 4. TILT is induced temporally in response to LPS stimulation in THP1 macrophages