Figures & data
Figure 1. Interaction between PCBP2 and mouse utrophin-A 5ʹUTR.
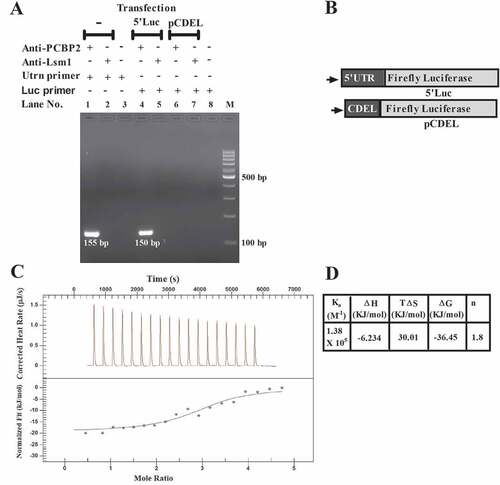
Figure 2. PCBP2 is a modulator of utrophin-A expression.
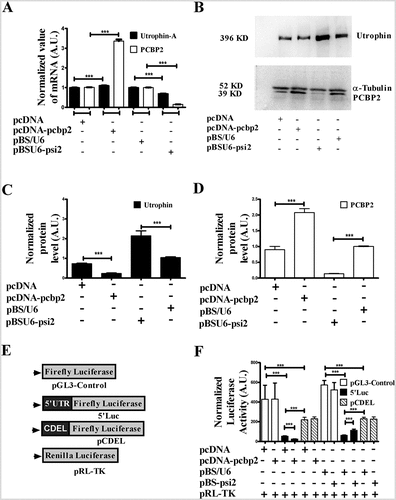
Figure 3. Interaction between PCBP2 and utrophin-A 5ʹUTR helps in the retention of utrophin-A transcript within the nucleus.
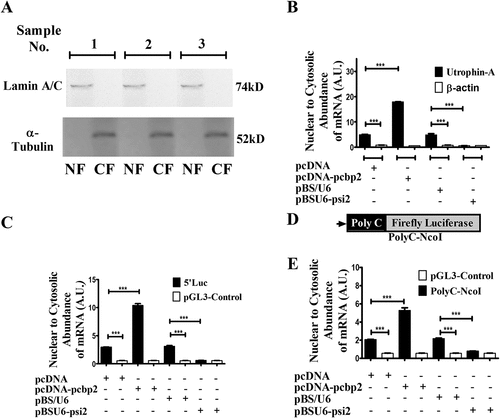
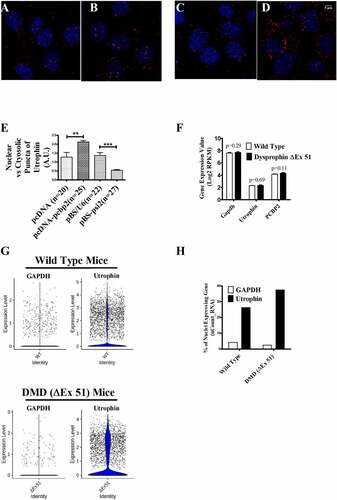
Figure 4. Localization of utrophin-A transcript in C2C12 cells and transcriptomic analysis of TA muscle from WT and DMD (ΔEx51) mice.