Figures & data
Figure 1. Progression of mRNA technology. The timeline illustrates the advancement of mRNA therapeutics, highlighting key milestones related to both general advancement of mRNA technology and the evolution of mRNA as a vaccine platform.
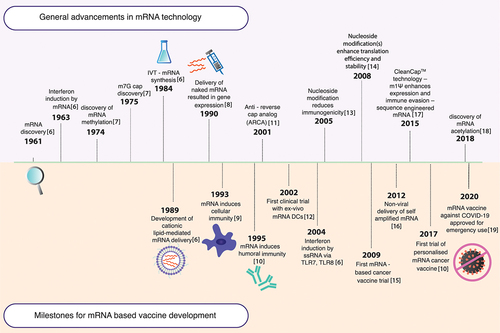
Figure 2. Activation of the immune system by mRNA vaccines. (1) The innate immune response is triggered via pathogen-associated molecular pattern (PAMP) recognition on non-self RNA. This recognition is mediated by pattern recognition receptors (PRRs) such as melanoma differentiation-associated 5 (MDA-5), nucleotide oligomerisation domain 2 (NOD2), and retinoic acid-inducible gene I (RIG-I) or toll-like receptors (TLRs), which ultimately results in type I interferon (IFN-I) release, allowing upregulation of proinflammatory genes, activation of the Th1 pathway and APC activation. (2) The internalised mRNA can be translated to generate the antigen, which is presented on the major histocompatibility complexes (MHC) on the APCs, initiating the adaptive response. (3) The antigen degraded by proteasomes is presented as peptides on the MHC I complex, leading to interactions with CD8+ cytotoxic T lymphocytes and triggering their maturation for a cellular response (indicated by blue arrows). (4) Additionally, antigens secreted exogenously can be internalised by other APCs, degraded into peptides within lysosomal endosomes, and presented on MHC II complexes on the cell surface (indicated by green arrows). This stimulates CD4+ T-helper cell interactions, which prime B lymphocytes on maturation for an antibody-mediated humoral response and generate memory cells specific to the target pathogen.
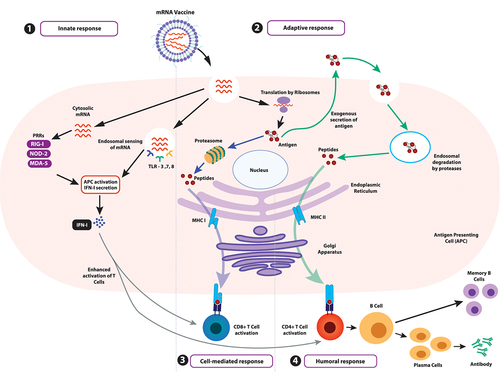
Figure 3. Synthesis of IVT-mRNA. (1) IVT-mRNA synthesis begins with a DNA template, usually pDNA, engineered to contain the gene of interest coding for the desired antigen identified by sequencing the genome of the target pathogen. (2) The pDNA template must include a bacteriophage promoter (T7), the ORF, a poly-deoxyribose T sequence (to code for the poly-A tail), and a restriction site which allows for restriction enzyme-mediated linearisation. (3) The linearised template undergoes in vitro transcription using a bacteriophage-derived T7 RNA polymerase. Following transcription, a mixture containing the desired mRNA, phage RNA polymerase, and nucleoside triphosphates is obtained. The 5’-cap can be introduced either enzymatically or be yielded in the transcription stage by including N7-methyl-guanosine analogue residues in excess of guanosine triphosphate (GTP) residues present. The 3’-tail can also be enzymatically added in this step, if not already incorporated. (4) The mixture is then purified using DNase I to degrade any contaminants and the template, followed by purification of the desired mRNA transcripts from a mix of abortive transcripts, longer transcripts with a 3’-overhang, oligodeoxynucleotides, and free nucleotides. While purification can be achieved by a series of precipitation and extraction steps, a chromatographic process is best suited for separating the transcripts of varied sizes. Techniques like high-performance liquid chromatography (HPLC) further refine the quality of the product, diminishing possibilities of unnecessarily activating innate immune sensors via contaminants. (5) Rapid mixing of the obtained mRNA with lipid via the utilisation of microfluidics allows for the self-assembly of mRNAs within lipid nanoparticles. (6) The obtained solution containing nanoparticles has to undergo further dialysis or filtration to remove any unencapsulated mRNA or non-aqueous solvent. Post filtration, the purified solution containing mRNA within lipid nanoparticles is ready for administration within the host.
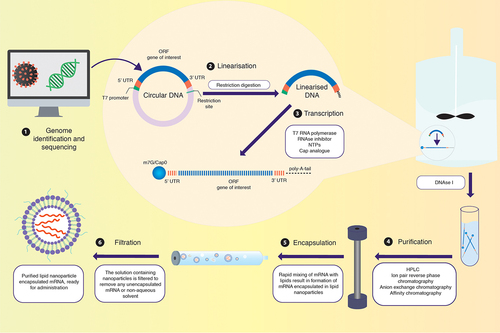
Figure 4. Strategy to optimise mRNA vaccines. mRNA can be modulated according to cell specificity in a number of ways: (1) By replacing nucleoside base(s) with chemically modified nucleoside(s), the translation efficiency and immunogenicity of mRNA can be altered. (2) Codon optimisation employed with GC-rich transcripts reduces immunogenicity to further increase translation efficiency and the safety profile of the mRNA. (3) By introducing stable 3’ UTR sequences, translation efficiency and stability can be modulated. (4) Manipulating the length of the 3’ poly(A) tail can improve mRNA stability. (5) Within the 5’ UTR, incorporation of GCC-(A/G)-CCAUGG sequence or avoidance of sequences similar to that of the ORF can enhance translation efficiency. (6) The 5’ cap promotes translation and further stabilises the mRNA. (7) The mRNA can be encapsulated within a variety of nanoparticle carriers for delivery into target tissues. This figure was created with BioRender.com.
Table 1. A systematic comparison of COVID-19 vaccines
Table 2. mRNA-based vaccines and therapeutics in clinical trials
