Figures & data
Figure 1. In-vitro synthesis of mt-tRNAMet assisted by cis-acting ribozymes. A ribozyme was placed either upstream of mt-tRNAMet as for (A) hammerhead (HH), (B) Hammerhead variant (HHV), (C) Pistol, and (D) Twister sister (TS), or downstream of mt-tRNAMet as for (E) twister. Sequences and secondary structures for each ribozyme are shown in blue on top of each panel. Sequences from tRNA-tRNAMet are shown in grey, and only the terminal ends are displayed. The black arrow indicates the cleavage site. Three independent IVT reactions were set up for each construct, and samples from different time points were resolved by denaturing gel electrophoresis. A typical experiment was shown at the bottom. Lanes 1–6 are samples taken at 1, 2, 3, 6, 12, and 24 hours of the transcription reaction. Lane Ctr is the control mt-tRNAMet in vitro transcribed without fusing with ribozymes. Pre stands for uncleaved precursor, P strands for the product mt-tRNAMet, and R stands for the freed ribozyme.
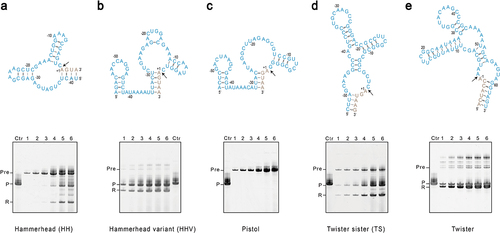
Figure 2. Characterization of sequence requirements at the catalytic sites of ribozymes. The constructs to produce mt-tRNAmet are the same as in , except that the conserved nucleotides at the catalytic sites are mutated. Sequences and secondary structures for (A) HHV, (B) Pistol, (C) TS, and (D) Twister are shown in blue on top of each panel. Conserved residues are indicated. Y: pyrimidine, R: purine. The black arrow indicates the cleavage site. For each ribozyme, various point or base-pair mutations were introduced into the conserved sites, and their effects on the ribozyme activity were tested. The IVT reactions of each construct at three hours were analysed by denaturing gel electrophoresis shown on the bottom panel. Lane Ctr is the control mt-tRNAMet in vitro transcribed without fusing with ribozymes. The label Pre stands for uncleaved precursor, P strands for the product mt-tRNAmet, and R stands for the cleaved ribozyme.
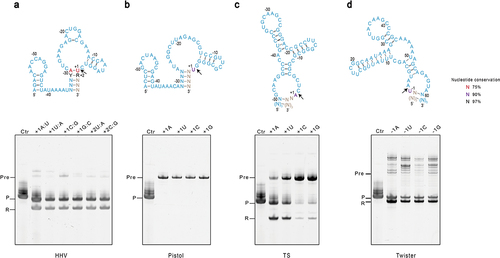
Figure 3. Engineering of ribozymes by introducing an affinity tag to the non-essential stem. A. Sequences of the three RNA affinity tags explored in this study: the MS2 stem-loop (MS2), the D8 aptamer (D8), and the K-turn motif (Kt). B. The affinity tag was attached to the non-essential stem of each ribozyme coloured in Orange. C and D. Activities of the engineered ribozymes. The modified ribozymes were used to produce mt-tRNAMet as in previous experiments, and RNA was analysed by denaturing gel electrophoresis. The red arrow indicates the position of the released mt-tRNAMet. The blue arrow points to the precursor, and the brown arrow points to the cleaved ribozymes.
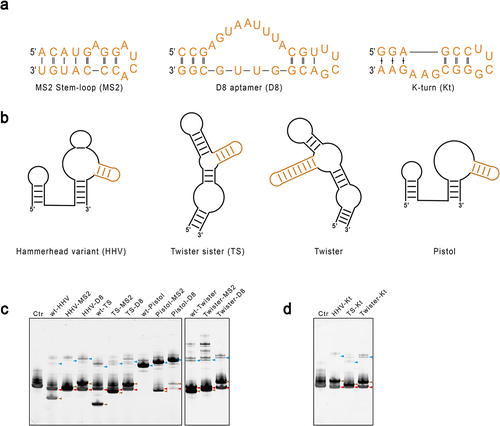
Figure 4. Post-transcriptional clean-up of IVT reactions using K-turn or MS2 affinity resins. A and B. K-turn modified ribozymes, HHV-Kt and Twister-Kt, were used to produce mt-tRNAMet with defined ends. C and D. MS2 modified ribozymes, HHV-MS2 and Twister-MS2, were used to produce mt-tRNAMet with defined ends. e-h. HHV-Kt and Twister-Kt were used in combination to produce the HCV IRES (E), the EMCV IRES (F), the Let-7 miRNA (G), and a 1KB RNA (H) with homogeneous 5’ and 3’ ends. Input is the IVT reaction before purification with the K-turn or MS2 affinity resin. FT is the flow-through of the resin. For HCV IRES, EMCV IRES, miRNA, and 1KB RNA, additional elution with binding buffer was performed (E1-E5). The RNA remaining on the resin was washed off with stripping buffer (W1-W5). Pre is the precursor transcript. R1 and R2 denote the Twister and HHV ribozymes, respectively. P is the released RNA product.
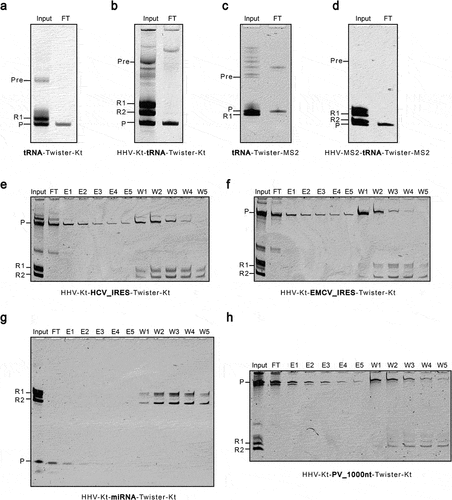
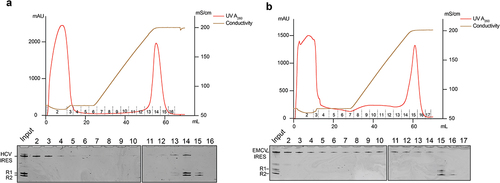
Figure 5. Large-scale production and purification of homogeneous HCV IRES and EMCV IRES with K-turn affinity resins. The ribozyme cassette HHV-Kt and Twister-Kt was used to produce HCV IRES (A) and EMCV IRES(B) with defined 5’ and 3’ ends. Following IVT, RNA was purified on a chromatography system with a prepacked K-turn affinity column. The chromatographic profile was shown on top of each panel. The bottom panel was the denaturing gel electrophoresis analysis of each fraction. R1 and R2 are cleaved Twister-Kt and HHV-Kt, respectively.