Figures & data
Figure 1. Schematic and predicted structure of vigilins. A) KH domain arrangement of two vigilins, the budding yeast Scp160 and the human HDLBP. Orange boxes represent classical KH domains, red ones represent diverged KH domains. B) COSMIC histogram indicating mutations across HDLBP in various cancer types. Mutations are depicted at an amino acid level correlating with HDLBP from a). Vertical lines display the location and relative frequency of substitutions C)Alphafold2 predicted protein structure of human HDLBP[Citation6,Citation7], using the same colouring scheme as in A).
![Figure 1. Schematic and predicted structure of vigilins. A) KH domain arrangement of two vigilins, the budding yeast Scp160 and the human HDLBP. Orange boxes represent classical KH domains, red ones represent diverged KH domains. B) COSMIC histogram indicating mutations across HDLBP in various cancer types. Mutations are depicted at an amino acid level correlating with HDLBP from a). Vertical lines display the location and relative frequency of substitutions C)Alphafold2 predicted protein structure of human HDLBP[Citation6,Citation7], using the same colouring scheme as in A).](/cms/asset/2522eed9-fef7-4ce5-bc37-d0d1e7110832/krnb_a_2313881_f0001_oc.jpg)
Figure 2. Architecture of a KH domain. A) definition of the minimal motif of a KH domain containing the GXXG loop between two α helices; B) comparison between classical (left) and diverged (right) KH domains in HDLBP (top) and the yeast vigilin Scp160 (bottom)[Citation6,Citation7].
![Figure 2. Architecture of a KH domain. A) definition of the minimal motif of a KH domain containing the GXXG loop between two α helices; B) comparison between classical (left) and diverged (right) KH domains in HDLBP (top) and the yeast vigilin Scp160 (bottom)[Citation6,Citation7].](/cms/asset/46d43f3f-2c5e-49db-a1b5-0002590baa37/krnb_a_2313881_f0002_oc.jpg)
Table 1. Summary of HDLBP-related cancer types and known interactors of HDLBP
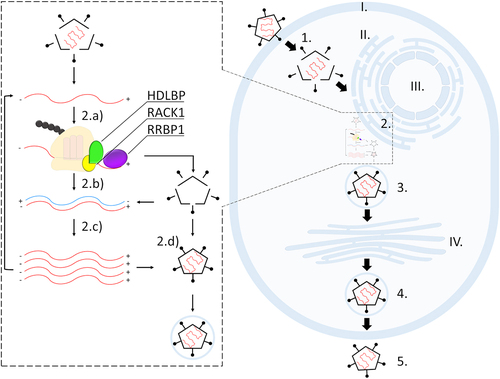
Figure 3. Simplified representation of a single-stranded positive-sense RNA virus infection; interaction of viral RNA (red) with HDLBP (green) during its translation at the ER. At the ribosome, HDLBP is bound to RACK1 (yellow) in proximity of RRBP1 (purple). Different compartments and steps during viral infection are indicated. I. plasma membrane; II. endoplasmic reticulum; III. nucleus; IV. Golgi apparatus. 1. Viral entry and disassembly; 2. Viral replication, including 2a. translation, 2b. transcription, 2c. viral RNA replication, and 2d. virion assembly.
Data availability statement
Data sharing not applicable – no new data generated.
