Figures & data
Figure 1. Flavivirus genome organization.
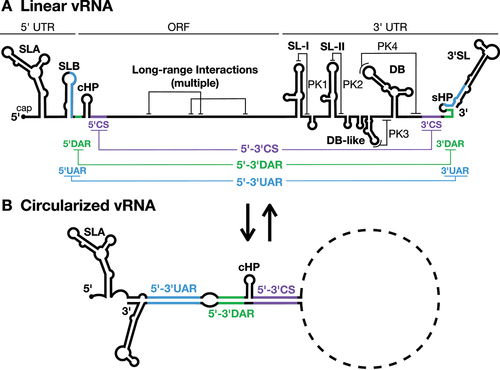
Table 1. RNA structures in the flavivirus genome and their roles in the viral life cycle.
Figure 2. The ‘L/V’ conformations of DENV and ZIKV SLA.
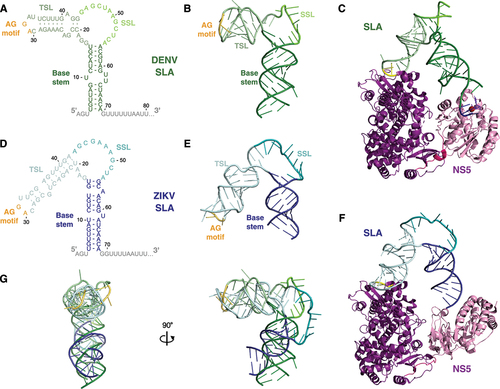
Figure 3. Models for the initiation of negative-strand RNA synthesis.
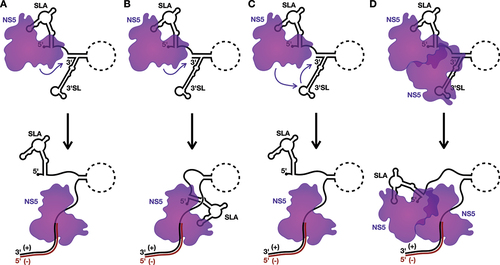
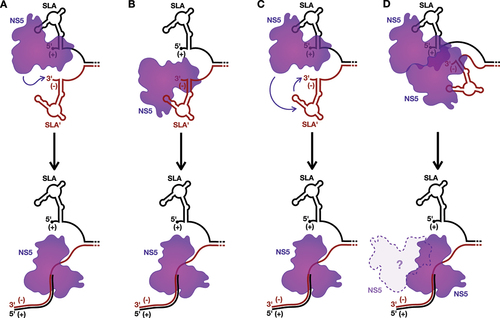
Figure 4. Models for the initiation of positive-strand RNA synthesis.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
