Figures & data
Figure 1. Subtle temperature dependent mRNA degradation decreases the number of functional transcripts.
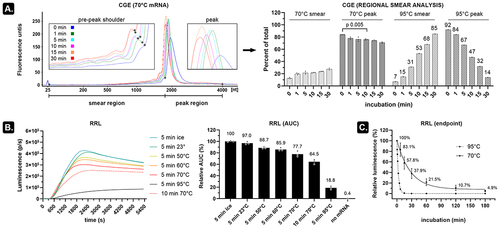
Figure 2. PNPase and oligo-T30 can be combined to selectively release measurable ADP from mRNA fragments.
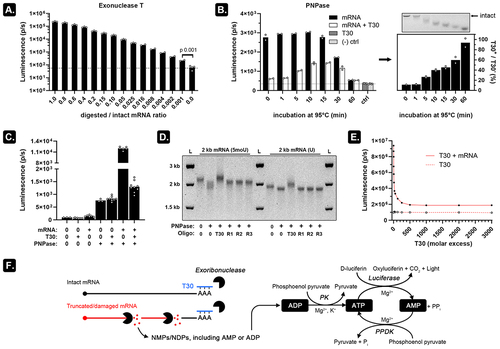
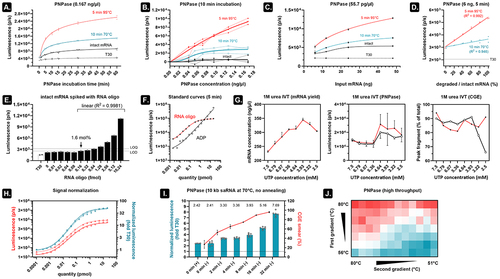
Figure 3. An optimized PNPase assay is fast, sensitive, and does not depend on the size of the mRNA analyte.
Combes2024_PNPaseAssay_supplement.docx
Download MS Word (270.1 KB)Figure Supplement.tif
Download TIFF Image (374 KB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
