Figures & data
Figure 2. Enzymatic activity and vacuolar localization of Atg15 are required for the maintenance of LD dynamics. (A) ATG15-deleted cells co-expressing GFP-tagged Atg15 and Vph1 tagged with mCherry (a vacuole marker) under their own promoter regulations were grown in SC medium for 48 h. The cells were harvested and subjected to fluorescence microscopy. The merged image consists of a green image representing the GFP signal and a red image for the mCherry signal. The corresponding lightfield (DIC) image is also shown. Scale bar: 5 µm. (B) Visualization of LDs in atg15 mutants by fluorescence microscopy. LDs were stained with BODIPY 493/503 after the cells were grown in SC medium for 48 h. Vector means the backbone plasmid for the expression of Atg15 variants (pRS316). The corresponding lightfield (DIC) images are also shown. Scale bar: 5 µm. (C) (Left columns) The relative amounts of TAG in the denoted strains after 48 h culture in SC medium were determined and shown so that the TAG level in the wild-type strain was presented as 100% and other values were normalized. The error bars indicate SD values. (Right columns) The numbers of LDs in the cells shown in (B) were counted and presented with SD values. At least 100 cells were used for the calculation. Statistical significance of the differences relative to the values of the atg15Δ strain harboring the vector was shown with asterisks (P< 0.01) and dagger symbols (P < 0.05).
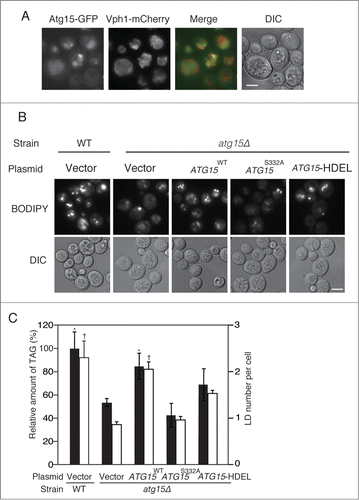
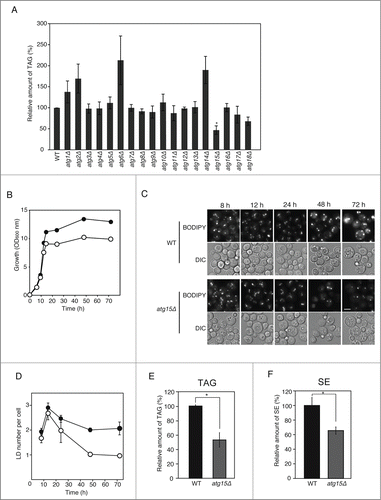
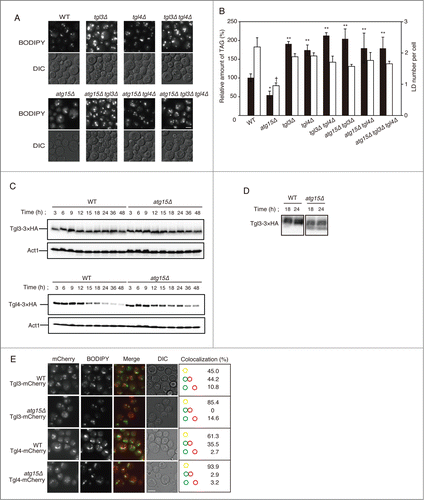
Figure 3. Effects of simultaneous disruptions of autophagy-related genes on LD abundance in the atg15Δ strain. (A) Schematic drawing of autophagosomes, autophagic bodies, and Atg15 in the denoted strains. (B) Fluorescence microscopy of LDs stained with BODIPY493/503 in the designated strains after culturing in SC medium for 48 h. The corresponding lightfield (DIC) images are also shown. Scale bar: 5 µm. (C) (Left columns) The relative amounts of TAG in the denoted strains after 48 h culture in SC medium were determined and shown so that the TAG level in the wild-type strain was presented as 100%. The error bars indicate SD values. (Right columns) The numbers of LDs in the cells shown in (B) were presented with SD values. At least 100 cells were used for the calculation. The asterisk and dagger symbols indicate P-values < 0.05, and the double asterisk indicates a P-value < 0.005, in comparison with the values of the atg15Δ strain. (D) Quantifications of LD numbers in various deletion mutants of autophagy-related genes. The numbers of LDs in the denoted strains cultured in SC medium for 48 h were counted after the staining of LDs with BODIPY493/503 and fluorescence microscopy. The error bars show SD values. At least 100 cells were used for the calculation. (E) Fluorescence microscopy of LDs in the denoted mutant strains stained with BODIPY 493/503 after culturing in SC medium for 48 h. The corresponding lightfield (DIC) images are also shown. Scale bar: 5 µm. (F) (Left columns) The relative amounts of TAG in the denoted strains after 48 h culturing in SC medium were determined and shown so that the TAG level in the wild-type strain was presented as 100% and the other values were normalized. The error bars indicate SD values. (Right columns) The numbers of LDs in the cells were counted and presented with SD values. At least 100 cells were used for the calculation. Statistical significance of the differences was shown with the asterisk and dagger symbols indicating P-values < 0.05, and the double asterisk indicating a P-value < 0.01.
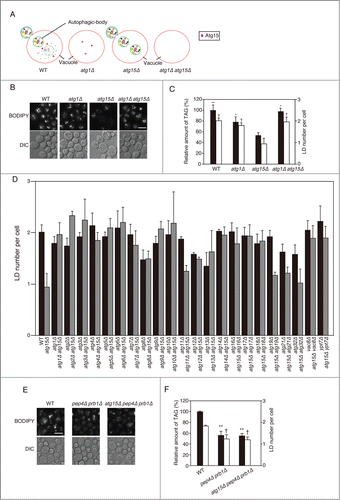
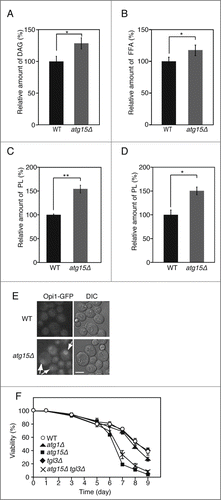
Figure 5. Increased levels of DAG, FFAs, and phospholipids in atg15Δ cells. (A) Quantification of DAG amount in wild-type and atg15Δ cells grown in SC medium for 48 h. The values from 3 independent experiments were plotted as mean ± SD. The asterisk indicates that the difference is statistically significant (P-value <0.05). (B) Quantification of FFA amounts in wild-type and atg15Δ cells grown in SC medium for 48 h. The values from 3 independent experiments were plotted as mean ± SD. The asterisk indicates that the difference is statistically significant (P-value <0.05). (C) Quantification of phospholipid amounts in wild-type and atg15Δ cells grown in SC medium for 48 h. The values from 3 independent experiments were plotted as mean ± SD. The double asterisk indicates that the difference is statistically significant (P-value <0.005). (D) Quantification of phospholipid amounts in microsome fractions of wild-type and atg15Δ cells grown in SC medium for 48 h. The values from 3 independent experiments were plotted as mean ± SD. The asterisk indicates that the difference is statistically significant (P-value <0.05). (E) Wild-type and atg15Δ cells expressing Opi1 tagged with GFP under its own promoter regulation were cultured in SC medium for 48 h. Cells were harvested and subjected to fluorescence microscopy. The GFP images along with the corresponding lightfield (DIC) images are shown. The arrows indicate Opi1-GFP signals exhibiting an ER-localization pattern. Scale bar: 5 µm. (F) Cell viabilities of the denoted strains cultured in SC medium for the indicated time periods were calculated by fluorescence microscopy of dead cells stained with phloxine B. The error bars represent SD values.