Figures & data
Figure 1. Expression of PEX5-KR triggers the removal of peroxisomes. SV40T-MEFs were transfected with plasmids encoding either (A) PEX5(L)-KR, (B) PEX5(L), or (C) KR. One day later, the cells were fixed, counterstained with DAPI, and processed for immunofluorescence with anti-PEX5 and/or anti-PEX14 antibodies followed by TxRed- and/or Alexa Fluor 488-conjugated secondary antibodies. (A) Upper and lower panels show a transfected cell where all, or most, peroxisomes are absent, respectively. Arrows indicate some of the remaining peroxisomes. Scale bar: 10 µm.
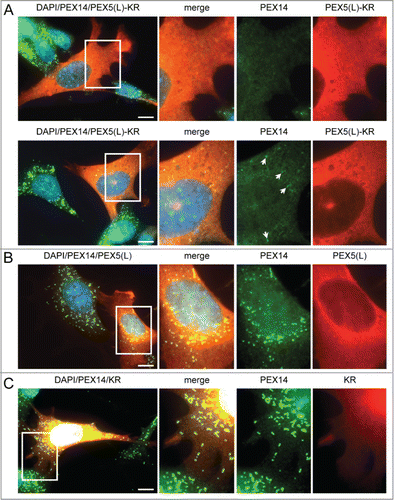
Figure 2. Expression of PEX5 proteins fused to a bulky C-terminal tag promotes a decrease in peroxisome number. SV40T-MEFs were cotransfected with plasmids encoding mitochondria-targeted EGFP (mt-EGFP; green color; marker for transfected cells) and either HsPEX5(S), HsPEX5(S)-FLAG, EGFP-HsPEX5(S), HsPEX5(S)-EGFP, HsPEX5(L), HsPEX5(L)-EGFP, HsPEX5(L)-KR, or mouse (Mus musculus, Mm) PEX5(L)-EGFP. One day later, the cells were fixed, counterstained with DAPI, and processed for immunofluorescence with anti-ABCD3 antibodies followed by TxRed- or Alexa Fluor 488-conjugated secondary antibodies. The number of peroxisomes in each transfected cell was counted and cataloged as more than 50 (>50 ), between 1 and 50 (1–50), or none (0). (A) Images of cells co-expressing mt-EGFP and PEX5(S)-EGFP with >50 (left panels), 1–50 (middle panels), or 0 (right panels) remaining peroxisomes are shown (these images depict representative examples of all phenotypes observed). Scale bar: 10 µm. (B) The percentage of transfected cells displaying each phenotype is plotted. The values above each bar represent the number of transfected cells analyzed per condition. A compilation of the results of at least 3 independent experiments (see Fig. S21) is shown. The “>50 peroxisomes” values from the “HsPEX5(S)” and “HsPEX5(L)” subpanels were statistically compared with the value from the corresponding control (−) condition (* p < 0.05; ** p < 0.01).
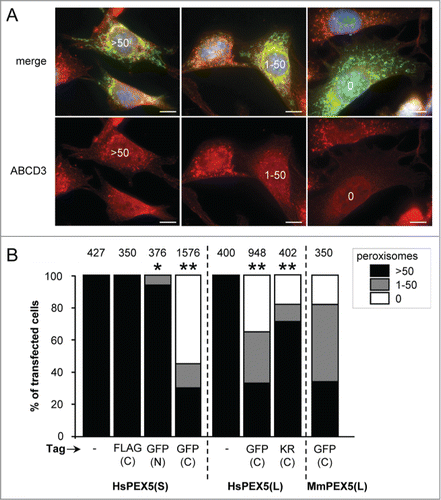
Figure 3. PEX5-EGFP-induced peroxisome removal is dependent on autophagy. Control (CT), Atg5+/+ or atg5−/− SV40T-MEFs were co-transfected with plasmids encoding mitochondria-targeted EGFP (marker for transfected cells) and a plasmid encoding either PEX5-EGFP or FLAG-SLC25A17-Ub in the absence (−) or presence of 10 mM 3-methyladenine (3-MA) or 10 µM LY294002 (LY). One day later, the cells were fixed and processed for immunofluorescence with either anti-ABCD3 or anti-PEX14 antibodies. Peroxisome degradation was quantified and plotted as in . The values above each bar represent the number of transfected cells analyzed per condition. (A, B) A compilation of the results of at least 3 independent experiments (see Fig. S22) is shown. The “>50 peroxisomes” values from the different (sub)panels were statistically compared with the value from the corresponding control condition (**, p < 0.01). (C) The results of a single experiment are shown.
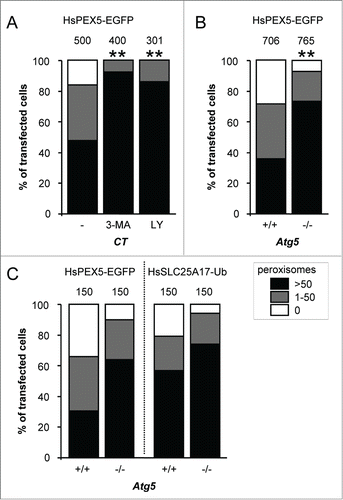
Figure 4. The N-terminal cysteine residue that marks PEX5 for recycling is crucial for PEX5-EGFP-induced pexophagy. SV40T-MEFs were cotransfected with plasmids encoding mitochondria-targeted EGFP (marker for transfected cells) and either (A) full-length PEX5(S)-EGFP (FL), PEX5(S)ΔN16-EGFP (ΔN16), or PEX5(S)ΔN110-EGFP (ΔN110), (B) PEX5(L) (FL), PEX5(L)C11K (C11K), PEX5(L)C11S (C11S), or PEX5(L)C11A (C11A), with or without a C-terminally fused EGFP-tag, or (C) PEX5(L)-EGFP (FL) PEX5(L)K527R-EGFP (K527R), PEX5(L)N526K-EGFP (N526K), or PEX5(S)ΔC299-PEX5(L)/PEX5RΔN326-EGFP (SWAP). For clarity reasons, the PEX5(L)-EGFP data are presented in panels (B and C). One day later, the cells were fixed and processed for immunofluorescence with anti-ABCD3 antibodies. Peroxisome degradation was quantified and plotted as in . The values above each bar represent the number of transfected cells analyzed per condition. A compilation of the results of at least 3 independent experiments (see Fig. S23) is shown. The “>50 peroxisomes” values from the (sub)panels were statistically compared with the value from the corresponding control (FL) condition (* p < 0.05; ** p < 0.01).
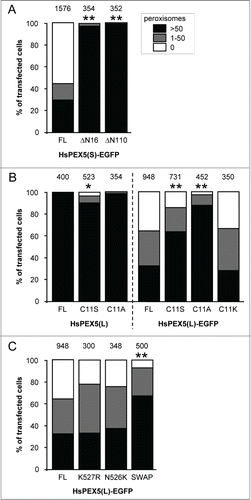
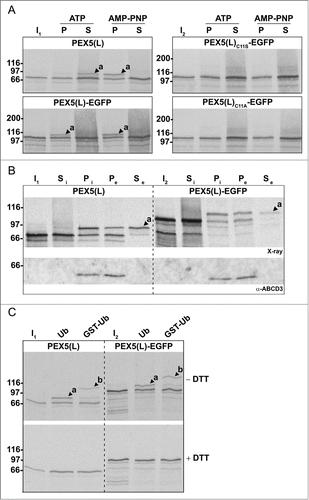
Figure 6. Monoubiquitinated PEX5-EGFP trapped at the DTM in the presence of ATP is only partially protease protected. Radiolabeled PEX5(L), PEX5(L)-EGFP and PEX5(L)C11A-EGFP were subjected to in vitro import assays in the presence of Ub aldehyde and either ATP or AMP-PNP, as indicated. After incubation at 37°C, organelle suspensions were treated with proteinase K. NEM-treated organelles were then isolated, and subjected to SDS-PAGE under reducing (+ DTT) and non-reducing (− DTT) conditions. The autoradiographs (upper panels) and a section of the corresponding Ponceau S-stained membranes (lower panels) are shown. a and b represent DTM-inserted PEX5(L) exposing 2 kDa of its N terminus to the cytosol and DTM-embedded monoubiquitinated PEX5(L), respectively. Citation21 The asterisks mark a set of PEX5(L)-EGFP-derived fragments that are protease resistant. Lanes I1, I2, I3, 5% of the radiolabeled protein used in the assays.
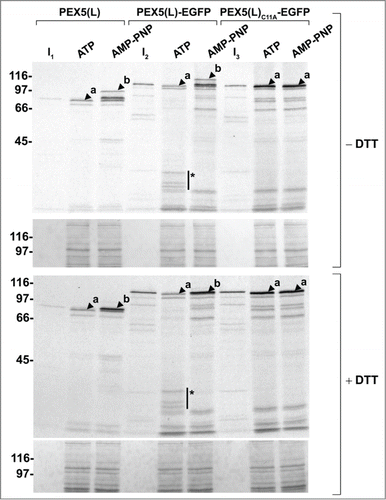
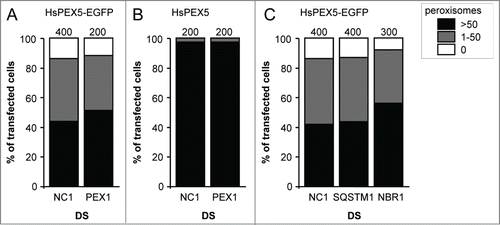
Figure 7. Downregulation of PEX1, SQSTM1, or NBR1 does not influence PEX5-EGFP-induced peroxisome removal. SV40T-MEFs were sequentially transfected with scrambled (NC1), PEX1-, SQSTM-, or NBR1-specific duplex siRNAs (DS) in combination or not with plasmids encoding mitochondria-targeted EGFP (marker for transfected cells) and (A, C) HsPEX5-EGFP or (B) HsPEX5 (for details, see Materials and Methods). One day later, the cells were fixed and processed for immunofluorescence microscopy with anti-PEX14 antibodies. Peroxisome degradation was quantified and plotted as in . The values above each bar represent the number of transfected cells analyzed per condition. A compilation of the results of at least 2 independent experiments (see Fig. S24) is shown. The “>50 peroxisomes” values from the (sub)panels were statistically compared with the value from the corresponding control (NC1) condition and found not to be statistically different.