Figures & data
Figure 1. Patterns of LC3B and HMGB1 immunohistochemical staining of breast adenocarcinomas in the training cohort. (A) LC3B puncta in a breast adenocarcinoma: nonmalignant breast gland (red arrow) staining negatively for LC3B, in the proximity of tumor cells with intense LC3B positivity. (B) Representative aspect of a breast adenocarcinoma without any detectable cytoplasmic LC3B puncta. (C) Cytoplasmic LC3B puncta in a breast adenocarcinoma that was considered as positive for LC3B staining. (D) Representative strong nuclear HMGB1 staining in normal mammary glands. (E) Representative aspect of a breast adenocarcinoma without any detectable nuclear staining (tumor considered as negative for nuclear HMGB1 expression). (F) Homogeneous nuclear HMGB1 staining in a breast adenocarcinoma considered as positive for HMGB1 nuclear expression.
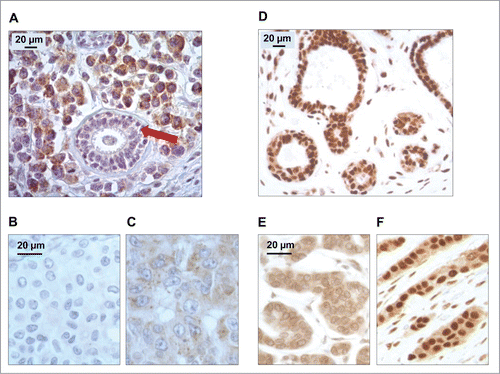
Figure 2. LC3B and HMGB1 staining quantitative analysis in the training cohort. (A) Distribution frequencies of percent of breast cancer cells harboring LC3B cytoplasmic puncta in the training cohort. (B) Distribution frequencies of percent of breast cancer cells with HMGB1-positive nuclei in the training cohort. (C) Correlation between percent of breast cancer cells with LC3B puncta, and percent of breast cancer cells with HMGB1 nuclear staining in each sample of the training cohort.
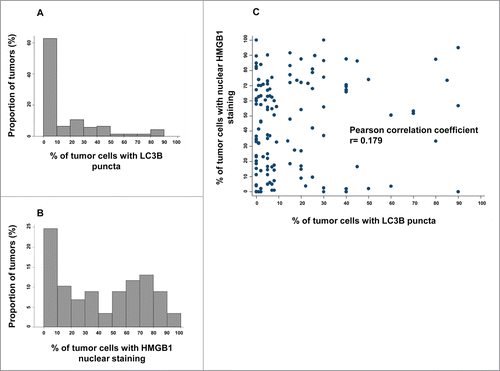
Figure 3. Kaplan-Meier survival plots of the training cohort. Metastasis-free survival (MFS) in the training cohort according to (A) cytoplasmic LC3B puncta in breast cancer cells (>10% = positive, or <10% = negative), (B) nuclear HMGB1 expression in breast cancer cells (>50% = positive, or <50% = negative). Breast cancer-specific survival (BCSS) in the PACS04 trial according to (B) cytoplasmic LC3B puncta and (D) nuclear HMGB1 expression. P values were calculated using the log-rank test.
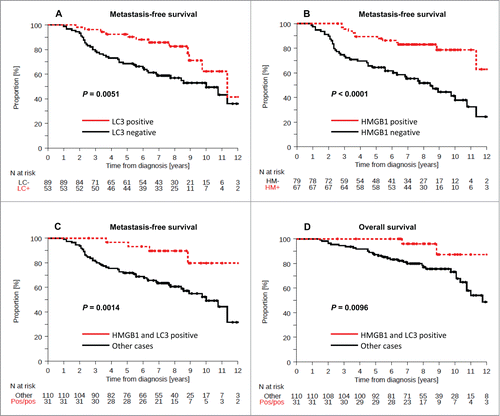
Table 1. Univariate and multivariate (incorporating LC3 or HMGB1) analysis (Cox regression) for factors associated with MFS in the training cohort
Figure 4. Kaplan-Meier survival plots for the validation cohort after analysis of LC3B and HMGB1. Metastasis-free survival (MFS) of BC patients included in the PACS04 trial after stratification of patients according to (A) cytoplasmic LC3B puncta in BC cells (>10% = positive, or<10% = negative), or (C) nuclear HMGB1 expression in breast cancer cells (>50% = positive, or <50% = negative). Breast cancer-specific survival (BCSS) in the PACS04 trial according to (B) cytoplasmic LC3B puncta and (D) nuclear HMGB1 expression. P values were calculated using the log-rank test.
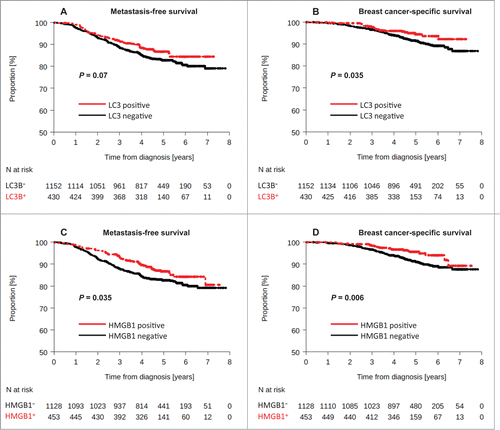
Table 2. Patients and tumor characteristics in PACS04 trial (N = 1581) according to HMGB1 and LC3 tumor status
Table 3. Multivariate analyses (Cox regression, including LC3 variable) for factors associated with MFS and BCSS in the validation cohort
Table 4. Univariate and multivariateFootnote* analysis (Cox regression) for factors associated with MFS
Figure 5. Kaplan-Meier curves for the validation cohort stratified after combined analysis of LC3B and HMGB1. Metastasis-free survival (A) and breast-cancer specific survival (B) in the PACS04 trial according to positivity for both cytoplasmic LC3B puncta and nuclear HMGB1expression in breast cancer cells. P values were calculated using the log-rank test.
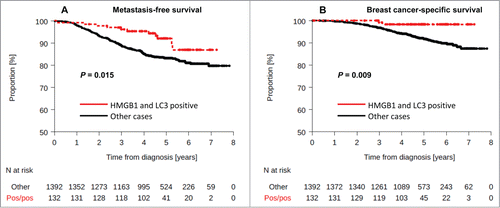
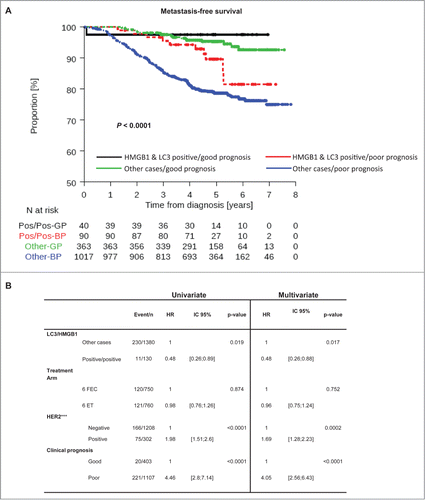
Figure 6. LC3B and HMGB1 double positivity as a complement to current prognostication tools. (A) Kaplan-Meier curves for metastasis-free survival (MFS) in the PACS04 trial stratified according to the presence of both cytoplasmic LC3B puncta and nuclear HMGB1 in breast cancer cells, as well as clinical prognostic status (good or poor). P values were calculated using the log-rank test. (B) Univariate and multivariate analyses (by Cox regression) of factors associated with MFS in the PACS04 trial. GP, good prognosis; BP, bad prognosis.