Figures & data
Figure 1. RAB24 localizes in LC3-positive vesicles. HeLa cells stably expressing myc-RAB24 were treated with a serum and amino acid-free medium (EBSS) for (A to C) 0, (D to F) 1, (G to I) 2 or (J to L) 4 h. Cells were labeled with RAB24 and LC3 antibodies and imaged with a confocal microscope. Inserts and arrows indicate vesicles positive for both proteins. See Figure S1 for quantitative analysis of the colocalization. Bar: 10 μm.
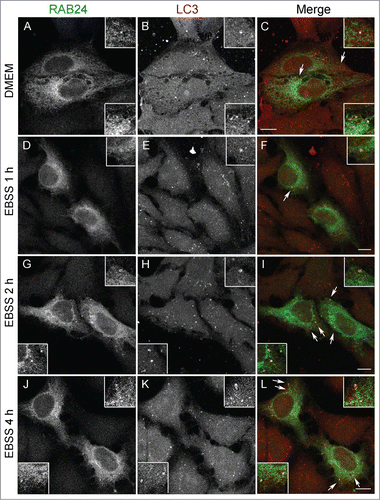
Figure 2. RAB24 localizes in the limiting membranes of autophagic compartments. HeLa (A i, B i, C) or NRK (A ii, B ii) cells were transfected with MYC-RAB24 and (A, C) left untreated in full-culture medium (DMEM), or (B) incubated in serum and amino acid-free EBSS for 90 min, or (C) incubated in EBSS with 100 nM Baf for 2 h. Cells were labeled with anti-RAB24 and anti-LC3 (A i and ii), or anti-RAB24 (B i and ii). (A i and ii) RAB24 label was in several cases observed to be ring-shaped, indicating it localized to the limiting membranes of the LC3-positive structures. (B i and ii) Immunoelectron microscopy confirmed the localization of RAB24 in the limiting membranes of autophagic compartments. Arrows indicate the outer limiting membrane and arrowheads the inner limiting membrane. (C) Subcellular fractionation of HeLa cells in a continuous OptiPrep gradient. The numbers above the western blot images indicate the 8 fractions collected from the top of the gradient. Highest OptiPrep concentration is in fraction 8 on the right. The fractions were detected for RAB24, LC3, SQSTM1 and transferrin receptor (TFRC). (D) Autolysosomal (AL) and dense lysosomal (L) membranes were isolated from rat liver and western blotting was used to detect RAB24, LC3, LAMP1 and RAB7 in these fractions. Bar in A i and ii: 10 μm. Gold particles in B i are 5 nm and ii 10 nm in diameter.
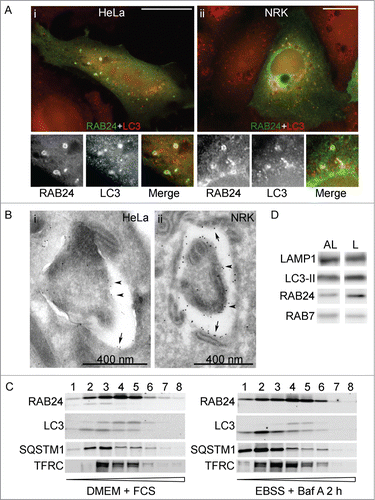
Figure 3. Analysis of RAB24 mutants. (A) GTP binding of RAB24S67L mutant was tested using GTP-agarose beads. Recombinant proteins were produced in E. Coli production strain BL21 and lysates were incubated with GTP-agarose gel. After washing, the bound protein was eluted from the gel with excess GTP, and western blotting was used to compare the eluted amounts of RAB24-WT and S67L. Eluted protein bands were normalized with their input bands. The columns and error bars show the mean and SEM from 4 independent experiments. (B) HeLa cells were transiently transfected with MYC-tagged wild-type or mutant MYC-RAB24 plasmids. Cell extracts were electrophoresed on urea (4 to 8 M) acrylamide (10–20%) SDS gradient gels and blotted on PVDF membranes. MYC antibody was used for detection. Prenylation-deficient mutants, CCΔ and CC→SS, lack the lower band, which represents the prenylated form of the protein. Nearly 40% of wild-type RAB24, and 45% of prenylation-competent mutants, are in the prenylated form. Tyrosine phosphorylation deficiency had a slight but nonsignificant effect on prenylation. The columns and error bars show the mean and SEM from 4 or 5 independent experiments. GFP-RAB7 is shown as a reference.
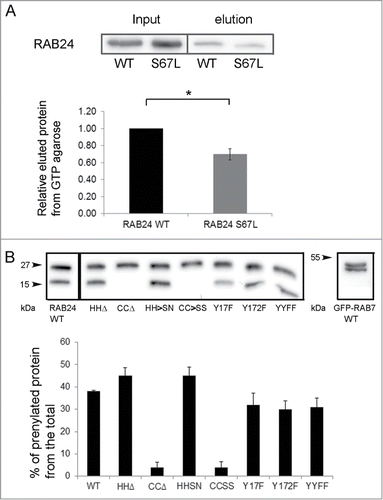
Figure 4. Guanine nucleotide binding is needed for the targeting of RAB24 to LC3 vesicles. WT or guanine nucleotide binding-deficient mutant RAB24S67L was expressed in NRK cells. (A to C and G to I) Cells were kept in full medium, DMEM, or (D to F and J to L) treated with a serum and amino acid-free medium, EBSS, for 4 h and after fixation labeled with antibodies against RAB24 and LC3. RAB24-WT localized to a perinuclear structure (arrowhead) and partially in LC3-positive puncta (arrows). RAB24S67L had a diffuse appearance and did not form the perinuclear staining pattern. See Figure S3 for quantification of the colocalization. Bar: 10 μm.
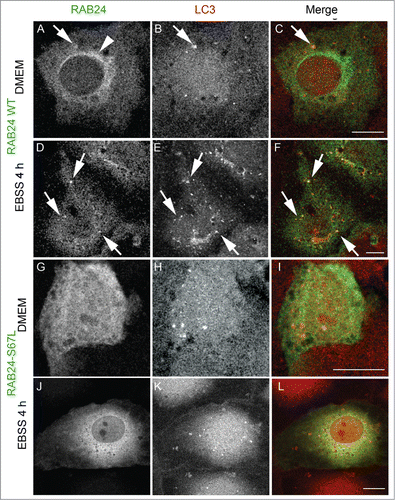
Figure 5. Prenylation-deficient mutants of RAB24 are not targeted to LC3 vesicles. Mutant RAB24 plasmids were expressed in NRK cells. Cells were kept in full-culture medium (DMEM) or treated with serum and amino acid-free medium (EBSS) for 4 h and after fixation labeled with antibodies against RAB24 and LC3. Prenylation-deficient mutants, CCΔ (A to F) and CCSS (G to L), had a diffuse appearance and did not form the perinuclear staining pattern observed for wild-type RAB24 (see ). Further, the RAB24 mutants did not localize in LC3 vesicles neither in normal medium nor in amino acid-free medium. For quantitative analysis of colocalization, see Figure S3. Bar: 10 μm.
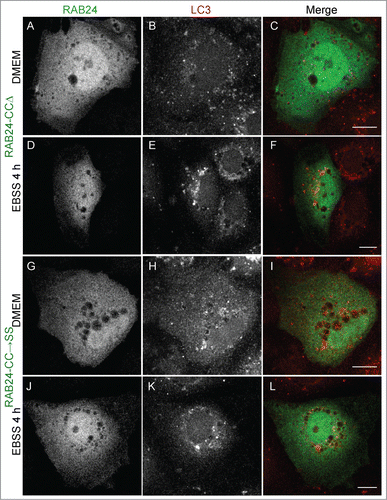
Figure 6. Prenylation-competent mutants of RAB24 are targeted to LC3 vesicles. Mutant RAB24 plasmids were expressed in NRK cells. Cells were kept in full-culture medium (DMEM) or treated with serum and amino acid-free medium (EBSS) for 4 h and after fixation labeled with antibodies against RAB24 and LC3. Prenylated mutants, HHΔ (A to F) and HHSN (G to L), resembled wild-type RAB24 with a perinuclear localization (arrowheads, compare with ). Further, both mutants partially colocalized with LC3 both in full culture-medium and after a 4 h EBSS treatment (arrows). Inserts highlight the colocalization in panels G to I. For quantitative analysis of colocalization, see Figure S3. Bar: 10 μm.
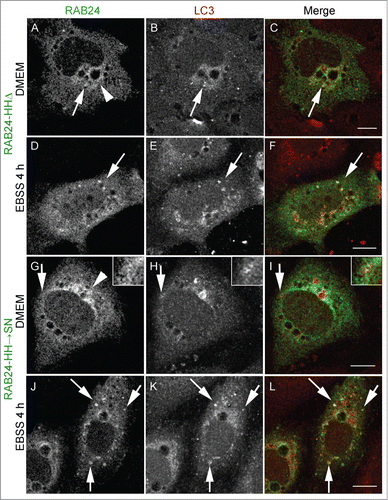
Figure 7. Tyrosine phosphorylation is less important for the targeting of RAB24 to LC3 vesicles. Mutant RAB24 plasmids were expressed in NRK cells. Cells were kept in full culture medium (DMEM) or treated with a serum and amino acid-free medium (EBSS) and after fixation labeled with antibodies against RAB24 and LC3. Tyrosine phosphorylation-deficient mutant YY17,172FF resembled wild-type RAB24 with perinuclear localization (arrowhead) (compare to ). RAB24 mutant partially colocalized with LC3 in full-culture medium (A to C, arrows) and after EBSS treatment (D to F, arrows). For quantitative analysis of colocalization, see Figure S3. Bar: 10 μm.
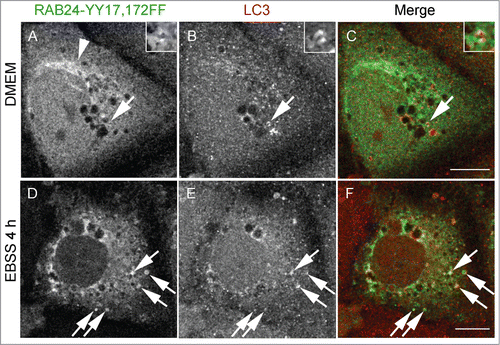
Figure 8. RAB24 is not needed for the formation or clearance of autophagosomes formed during 2-h serum and amino acid deprivation. (A) siRNA was used to silence RAB24 in HeLa cells and western blotting was used to monitor the protein levels. Tubulin is shown as a loading control. RAB24 silencing was approximately 95% as compared to 2 different siRNA controls, CTR, nontargeted siRNA and CTRRF, RISC-free control. (B) Quantitative electron microscopy was used to monitor the amount of autophagic compartments. The cells were either fixed without treatment (FCS), or incubated in serum and amino acid-free medium for 2 h (EBSS), or first incubated in EBSS for 2 h and then chased in full-culture medium for 2 h (EBSS-FCS), before fixation. (B and C) Immature (ACi) and degradative (ACd) autophagic compartments were quantified. Note that there was an increase in the amount of autophagic compartments in RAB24-silenced cells compared to control cells in full medium (FCS). For representative microscopy images, see Figure S5. The columns and error bars show the mean and SEM, respectively, from 3 to 4 counted grid squares, each containing a minimum of 50 to 60 cell profiles. Statistical significance was tested with Mann-Whitney U-test (P = 0.05). AC, autophagic compartment; ACd, degradative autophagic compartment; ACi, immature autophagic compartment.
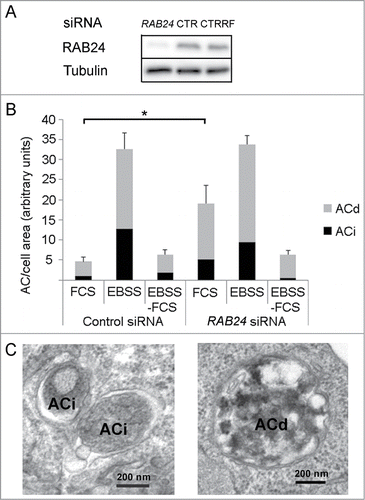
Figure 9. RAB24 is needed for the clearance of late autophagic compartments in full-culture medium conditions. Smart pool siRNA or single siRNA oligos (1, 2, or 4) from the pool were used to silence RAB24 in HeLa cells. The cells were either fixed without treatment, or incubated in fresh full culture medium for 2 h, or in fresh full-culture medium containing Baf (100 nM) for 2 h, before fixation. (A) Autophagic compartments were quantified by electron microscopy. Equal amounts of autophagic compartments were observed in the Baf-treated, RAB24-silenced and control cells. However, the RAB24 siRNA-transfected cells contained 4 times more autophagic compartments than the control cells in full-culture medium conditions. For representative image, see Figure S6. (A and B) The accumulating autophagic compartments in the RAB24-silenced cells were mostly degradative ACds. RAB24 silencing was 83% for the RAB24 smart pool and 55 to 69% for the single oligos. The columns and error bars show the mean and SEM, respectively, from a minimum of 40 images taken at 1500X primary magnification from 2 to 4 grid squares. The data are from one representative experiment out of 3 with similar results (P ≤ 3.341*10−6 for *** and P ≥ 0.618 for NS). AC, autophagic compartment; ACd, degradative autophagic compartment; ACi, immature autophagic compartment; NS, nonsignificant.
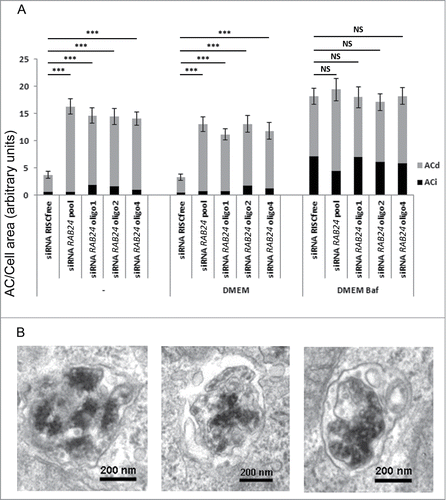
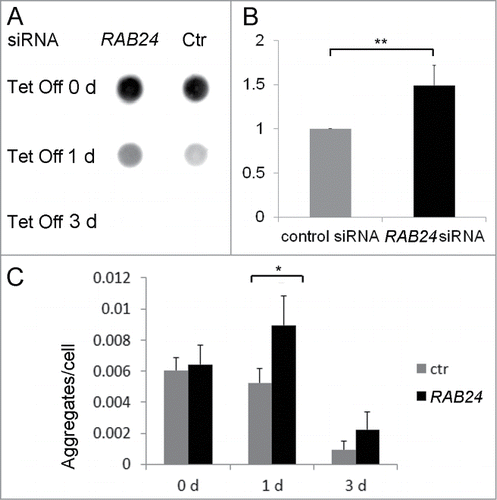
Figure 10. RAB24 facilitates the clearance of mutant HTT. siRNA was used to silence RAB24 in HeLa cells expressing HTT 65PQ. Two or 3 d after transfection with RAB24 or control siRNA, HTT mutant protein expression was inhibited by the addition of tetracycline (10 µg/ml) to the culture medium. The cells were given either 0, 1 or 3 d for aggregate clearance after which they were either treated for dot blot filter trap assay (A and B) or fixed and imaged by fluorescence microscopy (C). (B) Quantification of the intensity of one-d Tet Off dots on cellulose acetate membrane showed a slower aggregate clearance in RAB24-silenced cells compared to the control. The signals were negligible after 3 d in tetracycline and therefore were not quantitated. The columns and error bars show the mean and SEM from 4 independent experiments where the RAB24 silencing was 86% on average. (C) Quantitation of the HTT aggregates in fluorescence images revealed that there were more aggregates per cell in RAB24-silenced cells than in the control cells. The columns show the mean and SEM from 21 or 22 analyzed pictures, each containing several hundreds of cells (a total of 3637 to 18003 cells per sample). Representative images are presented in Figure S9. The experiment was repeated 2 times with similar results. The mean RAB24-silencing in the 2 experiments was 66% in 0-d samples and 67% in 3-d samples. Statistical significance was estimated using the Wilcoxon test for paired populations (P = 0.005).