Figures & data
Table 1. Statistics of ncRNA-associated cell death entries in ncRDeathDB
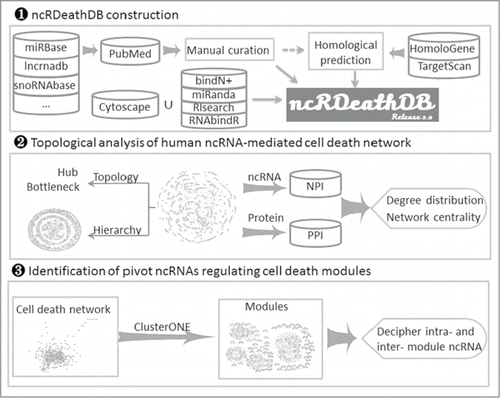
Figure 2. A flowchart for retrieving ncRNA-associated cell death interaction entry. (A) Three searching paths for retrieving the ncRNA-mediated cell death system. (B) The result of a representative database entry. (C) The detailed information for an ncRNA-associated cell death entry. In the results and detail pages, ncRDeathDB linked each ncRNA/protein to their corresponding detail pages and PMID to NCBI PubMed database.
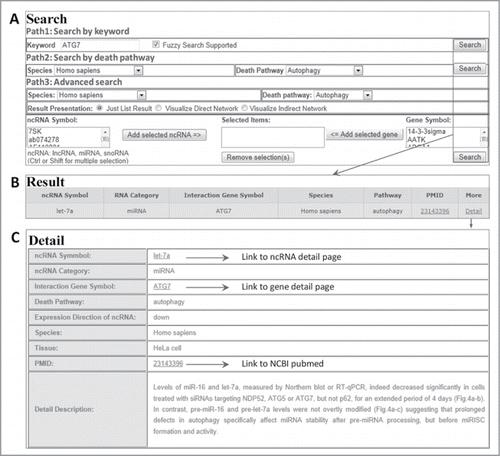
Figure 3. Schematic diagram for network measurements used to decipher the ncRNA-mediated cell death system. (A) A schematic diagram represents one bottleneck node, 2 hub nodes and one pivot node. Hubs are highly connected to other nodes in the network and act as the center point within the modules or subnetworks. Evidence from model organisms indicates that hub proteins tend to be encoded by essential genes, and the deletion of hub nodes leads to amount of diseases. Bottlenecks are defined as proteins with a high betweenness that are central to many paths in the network, and play a key role in controlling network signaling, and also like to be essential proteins, and often function as key genes in the development of disease progression. Pivots correspond to significantly shared members of 2 different pathways or subnetworks, which potentially have a role as molecular switches between these pathways or subnetworks, and further sheds light on the organization of the cellular machinery. (B) Schematic description of the 3-core decomposition of a network. k-core is a subnetwork defined by iteratively removing the nodes with a degree lower than k and their incident links. The inner k-cores were shown to be global hubs and enriched with lethal genes, which may potentially act as the evolutionarily conserved backbone of ncRNA-mediated cell death system.
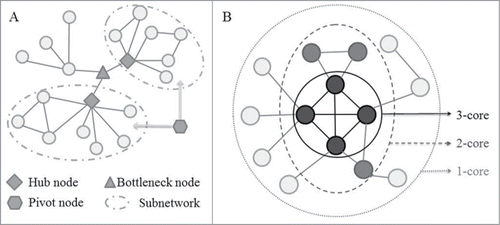
Figure 4. Distribution of degrees of apoptosis and autophagy-associated ncRNAs and proteins in the global human NPI and PPI network. The boxplot represents the degree distribution. The cell death-associated ncRNAs and proteins especially the dual nodes have a significantly higher degree distribution than that of the whole nodes in the NPI and PPI network.
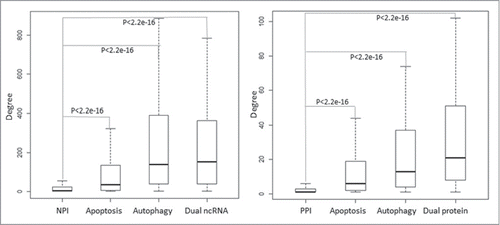
Figure 5. Analysis of network centricity of cell death-associated ncRNA and protein in global the human NPI and PPI network. The percent of both cell death-associated ncRNAs and proteins in k-cores of the NPI and PPI network significantly increased as k increased, and the dual proteins and ncRNAs involved in both autophagy and apoptosis increased the fastest.
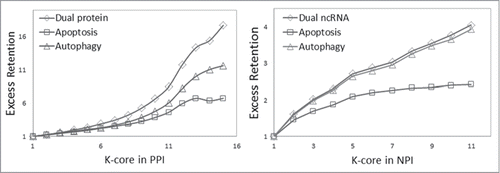
Table 2. ncRNAs are significant for the regulation of human protein autophagy subnetworks
Figure 6. Example of a module significantly regulated by ncRNAs identified in the human cell death protein interaction network. Pink and blue nodes represent apoptosis and autophagy, respectively. Yellow nodes represent the ncRNAs and proteins that play dual roles in apoptosis and autophagy. Triangular, arrowhead-shaped and circular nodes represent cell death-associated miRNAs, lncRNAs and proteins, respectively.
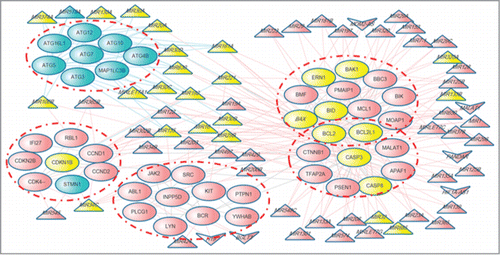
Table 3. Statistics of top 10 significant ncRNAs regulating human protein cell death modules