Figures & data
Figure 1. Correlations between DUSP1, LC3-II and SQSTM1 levels. Western blot analyses of DUSP1, LC3-I, LC3-II, ACTB or SQSTM1 (upper panel) and quantification of LC3-II (lower panel) in (A and D) CAOV3 cells stably transfected with DUSP1 shRNA (sh-DUSP1) or nontarget control shRNA (Nontarget), (B, E and G) Dusp1+/+ and dusp1−/− MEFs, and (C and F) TOV112D cells stably transfected with DUSP1 cDNA or empty vector. Cells (D-F) were left untreated or treated with 5 µM rapamycin (Rap) for 20 h prior to being harvested for western blot analyses. Cells (G) were left untreated or treated with 20 μM cisplatin for 24 h prior to being harvested for western blot analysis. Data represent mean±SD of 3 independent experiments. *, P < 0.01 and **, P < 0.005, statistically significant.
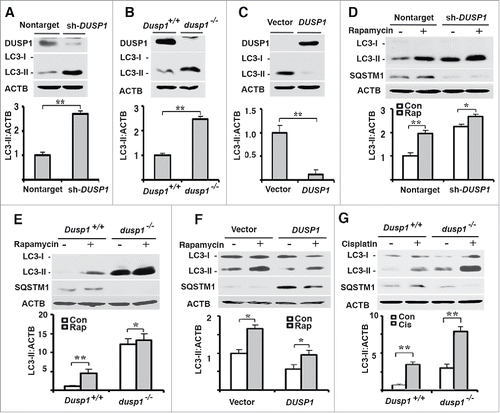
Figure 2. Inverse relationship between DUSP1 expression and autophagic vesicle formation. (A-C) Representative fluorescence images (upper panels) and quantification of GFP-LC3 puncta (lower panels) in (A) CAOV3 cells with DUSP1 shRNA (sh-DUSP1) or nontarget shRNA, (B) Dusp1+/+ and dusp1−/− MEFs and (C) TOV112D cells stably transfected with DUSP1 cDNA or empty vector. Cells were left untreated or treated with 5 µM rapamycin for 20 h prior to image capture. Bright puncta denote autophagosomal structures. (D) Representative indirect immunofluorescent images of endogenous LC3. DUSP1 shRNA or nontarget shRNA CAOV3 cells were left untreated or treated with 5µM rapamycin for 20 h prior to being processed for immunofluorescent detection of LC3. Red puncta denote autophagic vesicle structures (upper panel) and quantification of LC3 puncta (lower panel). Data represent mean ± SD of 3 independent experiments, in each of which 20 or more cells were counted. (E) Representative electron micrographs (upper panel) and quantification of autophagic vesicles (lower panel). DUSP1 shRNA or nontarget shRNA CAOV3 cells were left untreated or treated with rapamycin (5 µM, 20 h). Arrows denote autophagic vesicles. Nucl, nucleus. Scale bars: (A-D) 10 μm; (E) 1.7 µm (upper panels), 600 nm (lower panels). *, P < 0.01 and **, P < 0.005, statistically significant.
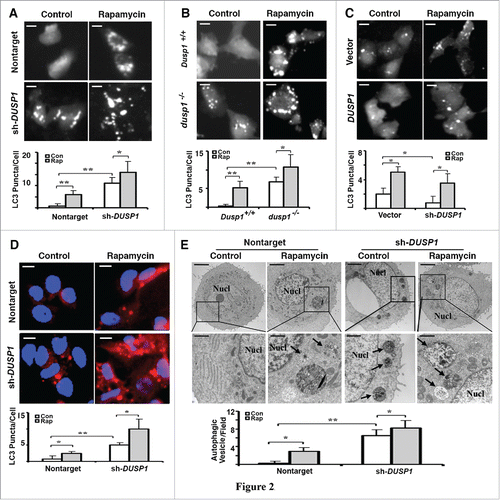
Figure 3. Effects of DUSP1 on autophagic flux. (A-D) Western blot analyses of DUSP1, LC3, SQSTM1 and ACTB (upper panel) and quantification of LC3 (lower panel). (A and D) DUSP1 shRNA (sh-DUSP1) or nontarget shRNA CAOV3 cells. (B) Dusp1+/+ and dusp1−/− MEFs. (C) Stable DUSP1 overexpression and empty vector control TOV112D cells. Cells were left untreated, or treated with 5 µM rapamycin and/or 10 nM bafilomycin A1 (A-C) or 10 μg/ml E64d (D) for 20 h before being harvested for western blot analyses. Data represent mean±SD of 3 independent experiments. *, P < 0.01 and **, P < 0.005, statistically significant.
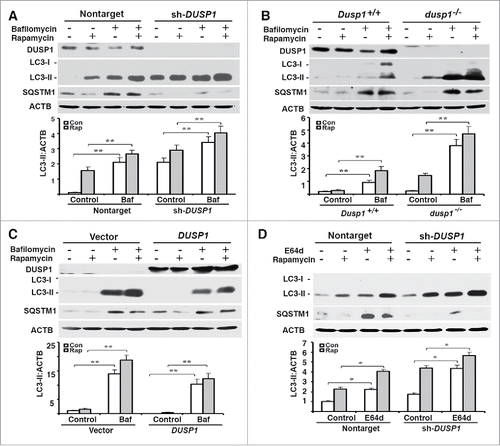
Figure 4. Effects of DUSP1 on MAPK activities and role of MAPKs in autophagy. (A) Western blot analyses of LC3, MAPK/p38, p-MAPK/p38, MAPK/ERK, p-MAPK/ERK, MAPK/JNK, and p-MAPK/JNK in control nontarget shRNA (nontarget) and DUSP1 shRNA (sh-DUSP1) CAOV3 cells (upper panel) and quantification of LC3 (lower panel). Cells were left untreated or treated with 5 µM rapamycin for 20 h prior to being harvested. (B) Western blot analyses of LC3, CREB, p-CREB, MAPK/ERK, p-MAPK/ERK, JUN and p-JUN in dusp1−/− MEFs treated with MAPK inhibitors (upper panel) and quantification of LC3 (lower panel). Cells were left untreated or treated with 10 µM SP600125 (SP), 10 µM U0126 (U) or 5 µM SB203580 (SB) for 24 h prior to being harvested for western blot analyses. (C) Western blot analyses of LC3, MAPK/ERK, and p- MAPK/ERK in nontarget and sh-DUSP1 CAOV3 cells treated with rapamycin (5 µM, 20 h), in the presence or absence of 10 µM U0126 (upper panel; LE, long exposure; SE, short exposure) and quantification of LC3 (lower panel). (D) Western blot analyses of MAPK/ERK and LC3 in sh-DUSP1 CAOV3 cells transfected with control siRNA (si-Control) or MAPK1-MAPK3 siRNA (si-MAPK1/3) (left panel) and quantification of LC3 (right panel). Cells were left untreated or treated with 5 μM rapamycin for 20 h prior to being harvested. (E) Dusp1+/+ and dusp1−/− MEFs were left untreated or treated with U0126 and then harvested for immunoprecipitation (IP) with antibody to BECN1. Immunoprecipitated proteins were analyzed by western blot with anti-BECN1, BECN1 phosphorylated at Ser15 (p-BECN1), phosphorylated MAPK/ERK and total MAPK/ERK. Whole cell lysates were used as input control. Data in (A-D) represent mean ± SD of 3 independent experiments. *, P < 0.01 and **, P < 0.005, statistically significant.
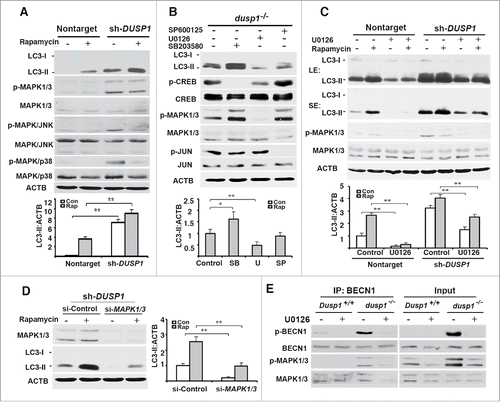
Figure 5. Effects of DUSP1 on RPS6KB, ULK1, and BECN1 phosphorylation. (A) Western blot analyses of the levels of total and phosphorylated RPS6KB (at Thr389) in Dusp1+/+ and dusp1−/− MEFs. Cells were left untreated or treated with 5 μM rapamycin for 20 h prior to being harvested. (B) Western blot analyses of the levels of p-ULK1 (at Ser555), ULK1, p-BECN1 (at Ser15) and BECN1 (upper panel), and quantification of p-ULK1 (Ser555) (middle panel) and p-BECN1 (Ser15) (lower panel). Dusp1+/+ and dusp1−/− MEFs were left untreated or treated with 5 μM rapamycin for 20 h prior to being harvested. (C) Western blot analyses of the levels of p-ULK1, ULK1, p-BECN1, BECN1 and MAPK1-MAPK3 in dusp1−/− MEFs (upper panel), and quantification of p-ULK1 (middle panel) and p-BECN1 (lower panel). Cells transfected with control siRNA (si-Control) or siRNA against Mapk1/3 (si-Mapk1/3) were left untreated or treated with 5 μM rapamycin for 20 h prior to being harvested. Data represent mean ± SD of 3 independent experiments. *, P < 0.01 and **, P < 0.005, statistically significant.
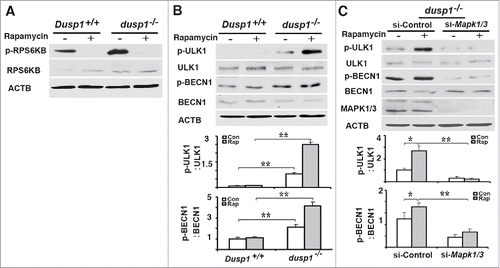
Figure 6. Effects of DUSP1 and MAPK/ERK on the association of PIK3C3, BECN1, ATG14, and p-MAPK/ERK. (A) Dusp1+/+ and dusp1−/− MEFs were left untreated or treated with 5 μM rapamycin for 20 h. Cell lysates were prepared, subjected to immunoprecipitation with antibody to PIK3C3, and then followed by western blot analyses using antibodies against BECN1, p-BECN1 (Ser15), ATG14 and p-MAPK1/ERK2-MAPK3/ERK1. Total cell lysates were used to assess input. (B) Dusp1−/− MEFs were transfected with control siRNA (si-Control) or siRNA against Mapk1-Mapk3 (si-Mapk1/3), and treatments as well as IP-western blot were performed as in (A). (C) Dusp1−/− MEFs were treated with 10 µM U0126 for 24 h prior to being processed for immunofluorescent detection of LC3. Red fluorescent puncta denote autophagic vesicle structures (upper panel) and were quantified (lower panel). (D) Dusp1+/+ MEFs transfected with pMCL-HA-Map2k1-R4F or control vector for 24 h were used for western blot analyses of HA, p-MAPK/ERK, MAPK/ERK, LC3 and ACTB (left panel) and immunofluorescent detection of LC3 (upper right panel). The lower right panel depicts quantification of red fluorescent puncta. The data in (C and D) represent means ± SD of analyses of minimally 20 cells. **, P < 0.005, statistically significant.
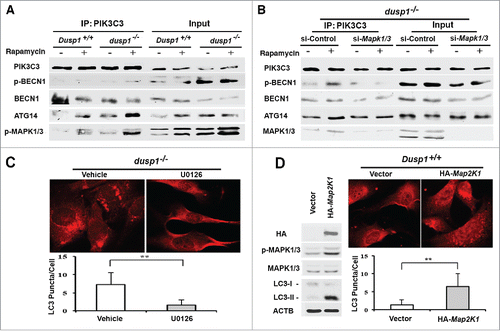
Figure 7. Cisplatin-resistant cells express high levels of DUSP1, low levels of activated MAPK/ERK and LC3-II, and decreased sensitivity to rapamycin. (A) CAOV3-P and CAOV3-CR cells were treated with different amounts of cisplatin for 48 h prior to being analyzed by MTT assays. (B) Western blot analyses of DUSP1 and MAPK/ERK in CAOV3-P and CAOV3-CR cells. Cells were left untreated or treated with 10 µM cisplatin for 20 h before being harvested. (C) Cisplatin-resistant cells have a reduced capacity for autophagy. CAOV3-P and CAOV3-CR cells were left untreated or treated with 5 µM rapamycin for 20 h prior to being harvested for western blot analyses of LC3 and SQSTM1 (upper panel) and quantification of LC3 (lower panel). (D) Cisplatin-resistant cells are less sensitive to rapamycin. CAOV3-P and CAOV3-CR cells were treated with the indicated doses of rapamycin for 48 h prior to MTT assays. Data represent mean ± SD of 3 independent experiments. *, P < 0.01 and **, P < 0.005, statistically significant.
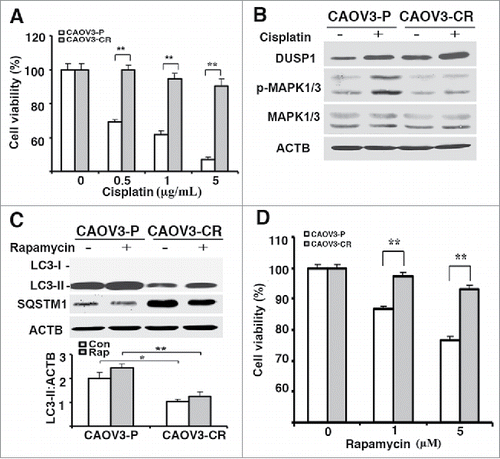
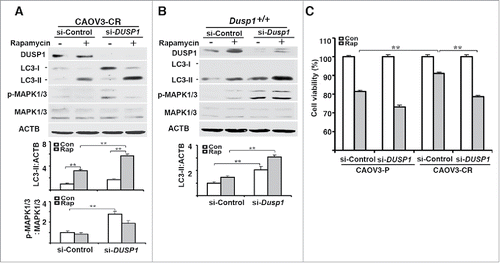
Figure 8. DUSP1-mediated suppression of autophagy is associated with cisplatin resistance in human ovarian cancer cells. (A) Effect of DUSP1 knockdown on LC3 level in cisplatin-resistant ovarian cancer cells. CAOV3-CR cells were transfected with control nontarget siRNA (si-Control) or siRNA against DUSP1 (si-DUSP1). Three days later the cells were either left alone or treated with 5 µM rapamycin for 20 h prior to being harvested for western blot analyses of DUSP1, MAPK/ERK, and LC3 (upper panel), quantifications of LC3 (middle panel) and p-MAPK/ERK (lower panel). (B) Effect of DUSP1 knockdown on LC3 level in MEFs. Dusp1+/+ MEFs were transfected with siRNA against Dusp1 (si-Dusp1) or control nontarget siRNA (si-Control) and treated with rapamycin as in (A). DUSP1, MAPK/ERK, and LC3 were determined by western blot analysis (upper panel) and LC3-II was quantified (lower panel). (C) Effect of DUSP1 on rapamycin sensitivity. CAOV3-P and CAOV3-CR cells were transfected with DUSP1 siRNA or control nontarget siRNA as in (A), and then left alone or treated with 5 μM rapamycin for 48 h prior to harvesting for MTT assays. Data represent mean ± SD of 3 independent experiments. **, P < 0.005, statistically significant.