Figures & data
Figure 1. PM2.5 exposure induced upregulation of VEGFA production in human bronchial epithelial cells. (A) Beas-2B cells were left untreated or were treated with different concentrations of PM2.5 (12.5, 25, 50, and 100 μg/mL) for 24 h. The production of VEGFA, IL1B, IL6, CXCL8 and TNF was then detected in the cell culture supernatant using ELISA (*, P < 0.05; **, P < 0.01). (B) Beas-2B cells were left untreated or were treated with PM2.5 as described in (A); then, the transcriptional responses of VEGFA, IL1B, IL6, CXCL8 and TNF were determined with an RT-PCR assay. (C) Beas-2B cells were transfected with a VEGFA promoter-driven luciferase reporter, and then stable transfectants were established. The transfectants were exposed to different doses of PM2.5 (as indicated), and the induction of VEGFA promoter-dependent luciferase activity was determined 12 h after PM2.5 exposure (**, P < 0.01). (D) Beas-2B cells were left untreated or were treated with PM2.5 as described in (A), and the intracellular VEGFA expression levels were examined using a western blot assay. (E) Beas-2B cells were left untreated or were treated with PM2.5 (100 μg/mL) for the indicated time periods; then, the intracellular VEGFA expression levels were measured. (F and G) Beas-2B cells were treated with PM2.5 (100 μg/mL) alone or in combination with PMB (50 μg/mL) for 24 h. Then, VEGFA transcription and protein synthesis and secretion were analyzed using RT-PCR, western blot and ELISA assays, respectively. (H) Primary human bronchial epithelial cells were left untreated or were treated with PM2.5 (20 µg/mL) for 24 h; then, VEGFA transcription and intracellular protein synthesis were detected using RT-PCR and western blot assays, respectively. (I) Primary human bronchial epithelial cells were left untreated or were treated with PM2.5 (20 μg/mL) for the indicated time periods; then, the production of VEGFA was detected in the cell culture supernatant using ELISA (**, P < 0.01).
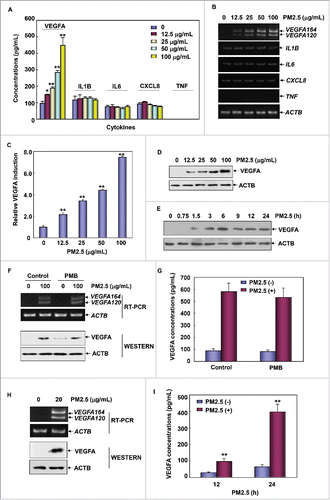
Figure 2. PM2.5 exposure induced autophagy in human bronchial epithelial cells. (A and B) Beas-2B cells were left untreated or were treated with PM2.5 as described in . Then, the expression levels of MAP1LC3B, BECN1 and SQSTM1 were examined. (C) Beas-2B cells were left untreated or were treated with PM2.5 (100 μg/mL) for 24 h; then, autophagy was examined under confocal microscopy after the cells were stained with Cyto-ID Green Autophagy Detection Reagent. (D) Beas-2B cells were treated with PM2.5 and were stained with Cyto-ID Autophagy Detection Reagent as described in (C). Then, the cells were collected and subjected to a flow cytometric analysis to quantitatively measure the autophagic fluorescence intensity inside the cells (**, P < 0.01). (E) TEM of Beas-2B cells untreated (left panel) or treated with 100 μg/mL of PM2.5 for 24 h (middle and right panels). The right panel shows a high-magnification image of the indicated region in the middle panel. Red arrows indicate double-membrane autophagic vesicles. Blue arrows indicate the particles taken up by the cells. (F) Beas-2B cells were treated with PM2.5 (100 μg/mL) alone or in combination with BafA1 (0.1 μM) during the final 4 h before the cells were harvested. Then, the expression levels of MAP1LC3B and SQSTM1 were examined 24 h after PM2.5 exposure. (G) Beas-2B cells were treated as described in , and the expression levels of MAP1LC3B, BECN1 and SQSTM1 were measured. (H) Beas-2B cells were treated as described in , and autophagy was examined under confocal microscopy after the cells were stained with Cyto-ID Green Autophagy Detection Reagent. (I) Primary human bronchial epithelial cells were left untreated or were treated with PM2.5 (20 μg/mL) for 24 h. Then, the expression levels of MAP1LC3B, BECN1 and SQSTM1 were measured. (J) Primary human bronchial epithelial cells were treated as described in (I) and autophagy was examined under confocal microscopy after the cells were stained with Cyto-ID Green Autophagy Detection Reagent.
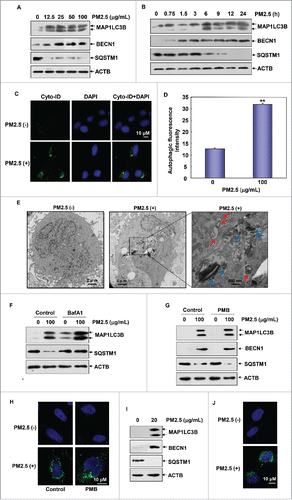
Figure 3. Autophagy induction was critical for mediating VEGFA production in Beas-2B cells upon PM2.5 exposure. (A) Beas-2B cells were pretreated with 3-MA, followed by exposure to PM2.5 (100 μg/mL). Then, the expression of VEGFA, BECN1 and MAP1LC3B was analyzed 24 h after PM2.5 exposure. (B) Beas-2B cells stably transfected with VEGFA promoter-driven luciferase reporter were treated with 3-MA and PM2.5 as described in (A). Then, the induction of VEGFA promoter-dependent luciferase activity was examined 12 h after PM2.5 exposure (**, P < 0.01). (C) Beas-2B cells were treated with 3-MA and PM2.5 as described in (A). Then, the production and secretion of VEGFA in Beas-2B cells were detected in the cell culture supernatant using ELISA 24 h after PM2.5 exposure (**, P < 0.01). (D) Beas-2B cells were transfected with ATG5 siRNA or control siRNA and then exposed to PM2.5 (100 μg/mL) 36 h after transfection. The expression of ATG5, VEGFA, BECN1 and MAP1LC3B was examined 24 h after PM2.5 exposure. (E) Beas-2B cells stably transfected with VEGFA promoter-driven luciferase reporter were transfected with ATG5 siRNA or control siRNA and treated with PM2.5 as described in (D). Then, the induction of VEGFA promoter-dependent luciferase activity was determined 12 h after PM2.5 exposure (**, P < 0.01). (F) Beas-2B cells were transfected and treated with PM2.5 as described in (D). Then, the production and secretion of VEGFA in Beas-2B cells were detected in the cell culture supernatant using ELISA 24 h after PM2.5 exposure (**, P < 0.01). (G) Beas-2B cells were transfected with BECN1 siRNA or control siRNA and then exposed to PM2.5 (100 μg/mL) 36 h after transfection. The expression of BECN1, MAP1LC3B and VEGFA was examined 24 h after PM2.5 exposure. (H) Beas-2B cells stably transfected with VEGFA promoter-driven luciferase reporter were transfected with BECN1 siRNA or control siRNA and treated with PM2.5 as described in (G). Then, the induction of VEGFA promoter-dependent luciferase activity was determined 12 h after PM2.5 exposure (**, P < 0.01). (I) Beas-2B cells were transfected and treated with PM2.5 as described in (G). Then, the production and secretion of VEGFA in Beas-2B cells were detected in the cell culture supernatant using ELISA 24 h after PM2.5 exposure (**, P < 0.01). (J) Beas-2B cells were treated as described in , and then VEGFA expression levels were measured 24 h after PM2.5 exposure. (K) Beas-2B cells stably transfected with VEGFA promoter-driven luciferase reporter were treated as in , and the induction of VEGFA promoter-dependent luciferase activity was examined 12 h after PM2.5 exposure (**, P < 0.01). (L) Beas-2B cells were treated as described in , and then the production and secretion of VEGFA in Beas-2B cells were detected in the cell culture supernatant using ELISA 24 h after PM2.5 exposure (**, P < 0.01).
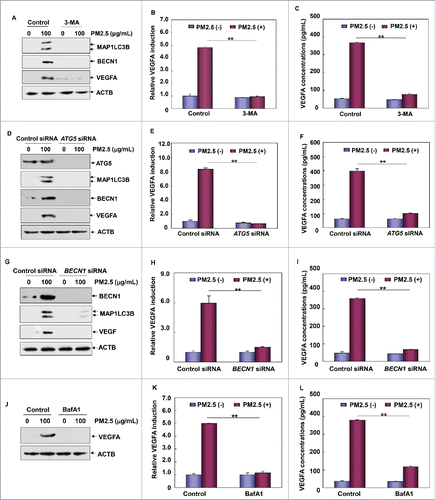
Figure 4. PM2.5-induced autophagy contributed to VEGFA production by activating the SRC-STAT3 pathway. (A) Beas-2B cells were left untreated or were treated with PM2.5 as described in ; then, the activation status of SRC and STAT3 was determined. (B) Beas-2B cells were transfected with STAT3-dependent luciferase reporter, and stable transfectants were established. The transfectants were exposed to different doses of PM2.5 (as indicated), and the induction of STAT3-dependent luciferase activity was examined 12 h after PM2.5 exposure (*, P < 0.05; **, P < 0.01). (C) Beas-2B cells were transfected with STAT3 siRNA or control siRNA; then, cells were treated with PM2.5 (100 μg/mL) 36 h after transfection. The expression of STAT3 and VEGFA was examined 24 h after PM2.5 exposure. (D) Beas-2B cells were transfected with SRC siRNA or control siRNA and then were treated with PM2.5 (100 μg/mL) 36 h after transfection. The expressions of SRC and VEGFA and the activation status of STAT3 was examined 24 h after PM2.5 exposure. (E) Beas-2B cells stably transfected with the STAT3-dependent luciferase reporter were transfected with SRC siRNA or control siRNA and exposed to PM2.5 (100 μg/mL) 36 h after transfection. The induction of the STAT3-dependent luciferase activity was determined 12 h after PM2.5 exposure (**, P < 0.01). (F) Beas-2B cells were treated as described in , and then the activation status of SRC and STAT3 was determined 24 h after PM2.5 exposure. (H, I and J) Beas-2B cells were treated or transfected as described in , 3C and 3E. Then, the activation status of SRC and STAT3 was determined 24 h after PM2.5 exposure. p, phosphorylated.
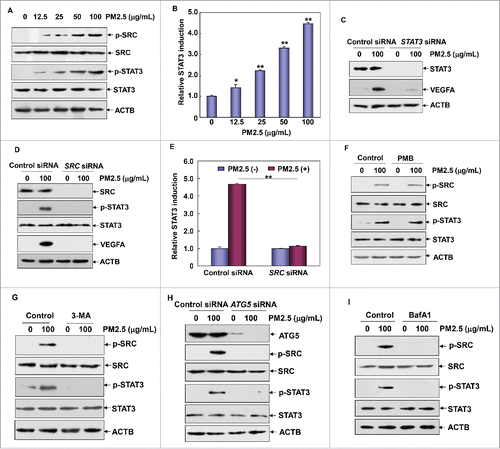
Figure 5. PM2.5 induced TP53 transactivation, which was critical for mediating autophagy induction in Beas-2B cells. (A and B) Beas-2B cells were left untreated or were treated with PM2.5 as described in , and the induction of TP53 activation and DRAM1 expression was examined. (C) Beas-2B cells were transfected with TP53-dependent luciferase reporter, and stable transfectants were established. The transfectants were exposed to different doses of PM2.5 (as indicated), and the induction of TP53-dependent luciferase activity was examined 12 h after PM2.5 exposure (**, P < 0.01). (D) Beas-2B cells were treated as described in , and the induction of TP53 activation and DRAM1 expression was examined 24 h after PM2.5 exposure. (E) Beas-2B cells were transfected with TP53 siRNA, DRAM1 siRNA or control siRNA; then, they were treated with PM2.5 (100 μg/mL) 36 h after transfection. The activation status of TP53 and the expression levels of DRAM1, BECN1 MAP1LC3B were examined 24 h after PM2.5 exposure. (F and G) Beas-2B cells were transfected and treated as described in (E), and then the autophagy signals were detected as described in (**, P < 0.01). p, phosphorylated.
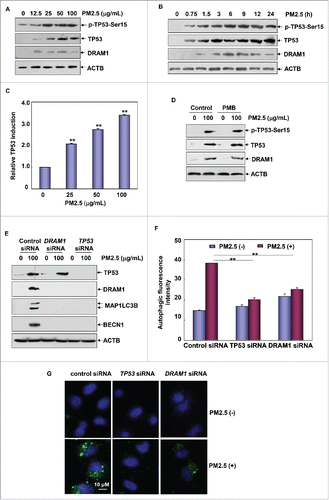
Figure 6. ATR was required for the induction of TP53-dependent autophagy in Beas-2B cells upon PM2.5 exposure. (A) Beas-2B cells were treated as described in , and then the activation status of ATM and ATR was determined. (B) Beas-2B cells were transfected with ATR siRNA, ATM siRNA or control siRNA; then, cells were treated with PM2.5 (100 μg/mL) 36 h after transfection. The activation status of TP53 and the expression levels of ATR, ATM, DRAM1, BECN1 and MAP1LC3B were examined 24 h after PM2.5 exposure. (C) Beas-2B cells stably transfected with TP53-dependent luciferase reporter were transfected with ATR siRNA, ATM siRNA or control siRNA; then exposed to PM2.5 (100 μg/mL) 36 h after transfection. The induction of TP53-dependent luciferase activity was determined 12 h after PM2.5 exposure (**, P < 0.01). (D and E) Beas-2B cells were transfected with ATR siRNA or control siRNA and then treated with PM2.5 (100 μg/mL) 36 h after transfection. The autophagy signals were detected as described in (**, P < 0.01). p, phosphorylated.
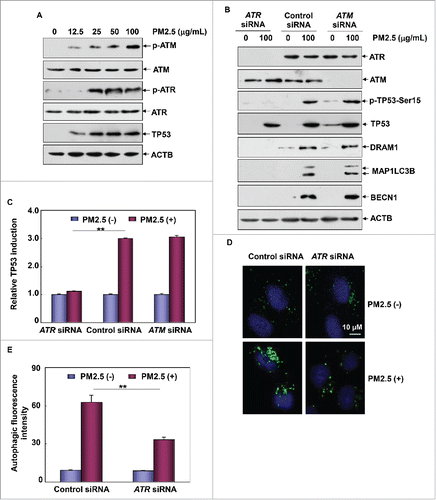
Figure 7. CHEK1 was the downstream target of ATR that mediated TP53-dependent autophagy in Beas-2B cells under PM2.5 exposure. (A) Beas-2B cells were treated as described in , and the activation status of CHEK1 was determined. (B) Beas-2B cells were transfected with ATR siRNA or control siRNA and treated with PM2.5 (100 μg/mL) 36 h after transfection. The activation status of CHEK1 was determined 24 h after PM2.5 exposure. (C) Beas-2B cells were treated as described in , and the activation status of the ATR-CHEK1 axis was determined 24 h after PM2.5 exposure. (D) Beas-2B cells were transfected with CHEK1 siRNA or control siRNA and then treated with PM2.5 (100 μg/mL) 36 h after transfection. The activation status of CHEK1 and TP53 and the expression levels of DRAM1, BECN1 and MAP1LC3B were examined 24 h after PM2.5 exposure. (E) Beas-2B cells stably transfected with TP53-dependent luciferase reporter were transfected with CHEK1 siRNA or control siRNA and exposed to PM2.5 (100 μg/mL) 36 h after transfection. The induction of TP53-dependent luciferase activity was determined 12 h after PM2.5 exposure (**, P < 0.01). (F and G) Beas-2B cells were transfected with CHEK1 siRNA or control siRNA and treated with PM2.5 (100 μg/mL) 36 h after transfection. Then, the autophagy signals were detected as described in (**, P < 0.01). p, phosphorylated.
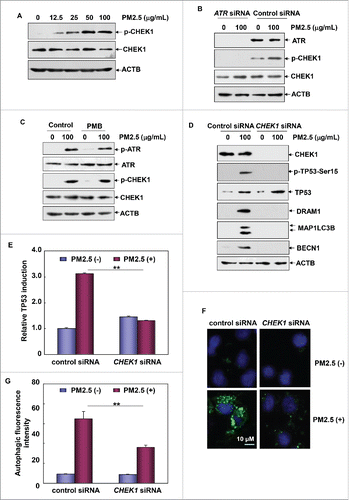
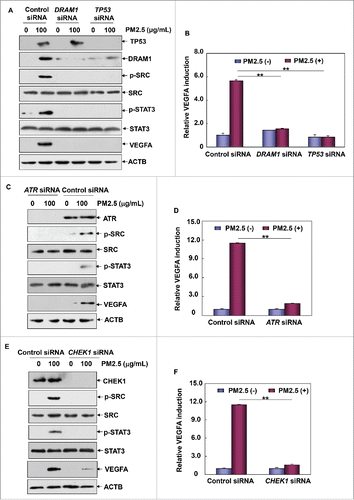
Figure 8. ATR-CHEK1-TP53 signaling pathway activation was required for mediating autophagy-dependent VEGFA production in Beas-2B cells under PM2.5 exposure. (A, C and E) Beas-2B cells were transfected with TP53 siRNA or DRAM1 siRNA (A), ATR siRNA (C) or CHEK1 siRNA (E) and their respective control siRNAs; then, cells were treated with PM2.5 (100 μg/mL) 36 h after transfection. The activation status of SRC and STAT3 and the expression level of VEGFA were determined 24 h after PM2.5 exposure. (B, D and F) Beas-2B cells stably transfected with a VEGFA promoter-driven luciferase reporter were transfected and treated as described in (A, C and E), respectively. Then, the induction of VEGFA promoter-dependent luciferase activity was determined 12 h after PM2.5 exposure (**, P < 0.01). p, phosphorylated.