Figures & data
Figure 1. Screenshot of an iLIR database data page. In the ‘LIRCPs’ menu, the user can access the full data available in the database for each model organism. The data are arranged in a table giving various information for each entry, such as the Uniprot Accession ID and protein name, the position and sequence of the xLIR as well as the position-specific scoring matrix (PSSM) score and the similarity of other validated LIR motifs. The data can be downloaded directly.
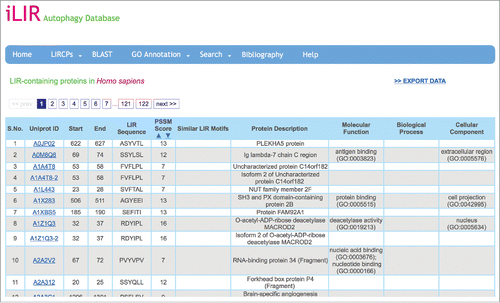
Figure 2. Screenshot of the iLIR database BLAST results page. Using the ‘BLAST’ menu, the user has the possibility to blast the sequence of interest against the sequences for one or all organisms available in the database in order to identify similar putative LIRCPs. The results page gives the list of similar sequences and the position of the putative LIR motif is indicated with red asterisks. The ‘sp’ and ‘tr’ preceding the FASTA header of the sequences producing a significant alignment refer to UniProtKG/Swiss-Prot (reviewed and manually annotated sequences) and UniProtKG/TrEMBL (unreviewed, automatically annotated sequences from large-scale screens), respectively.
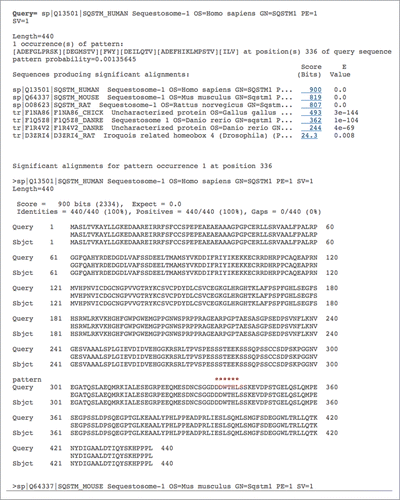
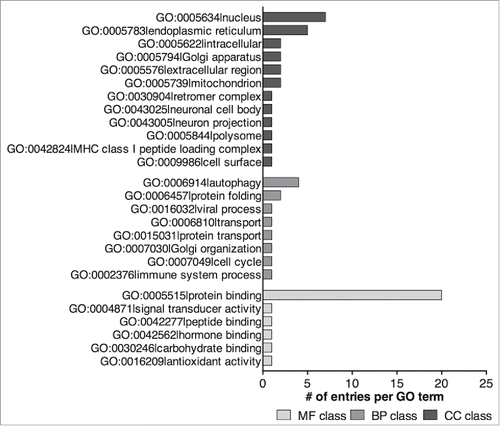
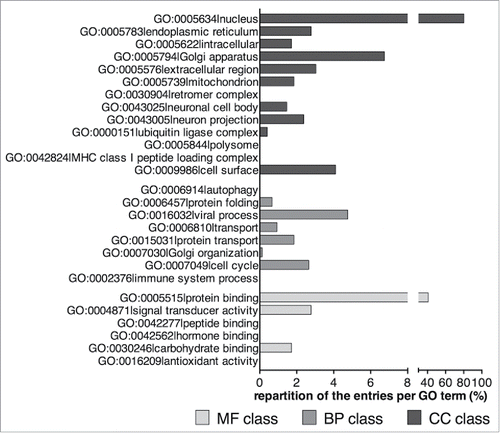
Figure 3. Representation of the number of ORFs (bar chart, plotted on the left axis) and percentage of putative LIRCPs identified (dot chart, plotted on the right axis) for each of the model organisms analyzed.
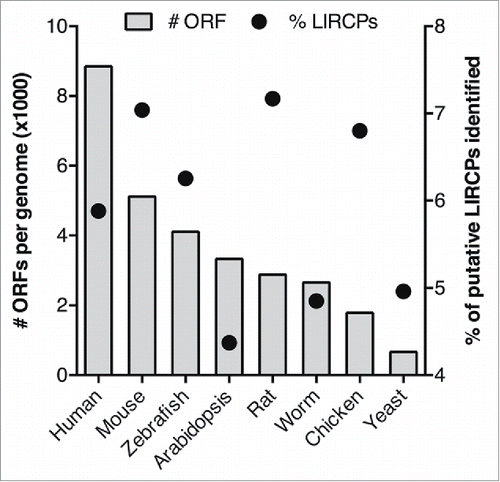
Table 1. Summary of the numbers of putative LIRCPs and LIR identified for the 8 model organisms studied.