Figures & data
Figure 1. NUPR1 depletion induces autolysosomal vacuolization. (A) IHC staining with anti-NUPR1 was performed on 118 NSCLC samples and their adjacent tissues. Representative images show moderate (case #1) and strong (case #2) NUPR1 staining. Scale bars: 10 µm. (B) Kaplan-Meier overall survival rates for 118 NSCLC subjects with low (0 to 5.0 staining scores, blue lines; n = 68) versus high (5.1 to 10.0 staining scores, green lines; n = 50) NUPR1 expression. Median survival was more than 80 mo for the low NUPR1 expression group versus 28 mo for the high NUPR1 expression group (P = 0.00025). (C and D) Relative NUPR1 transcript levels determined by quantitative RT-PCR shown as fold differences relative to GAPDH in a normal lung epithelial cell line (NHBE) and cancer cell lines as indicated in (C), and the NUPR1 level determined by western blotting is shown with ACTB as a loading control in (D). (E) Western blot confirming the knockdown efficiency of 3 shRNAs against human NUPR1, with fire fly luciferase as a negative control (con) and GAPDH as an internal control. (F) Representative phase-contrast micrographs of cell morphological changes following the expression of NUPR1 shRNA in A549 cells. Large and small vacuoles can be seen scattered throughout the cytoplasm in NUPR1-knockdown cells (arrows). The right graph shows the quantification of the number of vacuoles per cell from 3 different experiments (mean ± SEM). Lower panels show transmission electron micrographs (TEM) images of A549 control and A549-NUPR1 shRNA cells at the indicated magnifications. NUPR1 depletion leads to accumulation of dilated autolysosomes (arrows). The right image is a higher magnification of the indicated portion, showing electron-dense material within autolysosomes. (G) Light micrographs and electron micrographs of cell morphology following NUPR1 depletion in H1299, H460 and H1155 cells. Arrows show the vacuole membrane location.
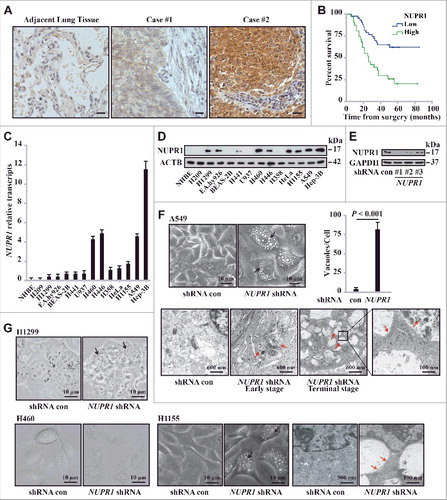
Figure 2. NUPR1 regulates autophagic flux and autolysosomal efflux. (A) Representative fluorescence images of A549 cells transiently expressing GFP-LC3B, with NUPR1 knockdown as described in the Materials and Methods. Arrows indicate autolysosomes, whereas arrowheads indicate autophagosomes. Bar graph on the right shows the number of GFP-LC3 puncta and vacuoles per cell. Scale bars: 5 µm. (B) Representative images adapted from time-lapse movies of A549-mCherry-GFP-LC3 cells treated with the indicated shRNA. Autophagosomes (arrowheads), yellow puncta; autolysosomes (arrows), red-only puncta. Quantification of the number of LC3 puncta per cell in NUPR1-depleted and control A549 cells (10 cells per group). Scale bars: 10 µm. (C) Immunoblot analysis of LC3B and SQSTM1 levels in A549 and H460 cells expressing shRNAs against NUPR1 used in E, with ACTB/β-actin as a loading control. (D) Cellular morphology after sequential knockdown of ATG5 and/or NUPR1 in A549 cells. The right bar graph shows the number of vacuoles per cell. Error bars represent the SD (n = 10). Scale bars: 10 µm. (E) Immunoblot analysis was performed as in (C) by knockdown of ATG5 and/or NUPR1. The left upper panel shows ATG5 knockdown efficiency. Graph depicts densitometric analysis of protein intensity as indicated, normalized to ACTB levels and expressed as fold change from untreated control (right panel). (F) Representative morphological changes in cellular vacuolization by NUPR1 depletion with or without 5 nM BafA1 treatment as indicated. The right panel is the quantification of the vacuoles per cell shown in the left panel. Scale bars: 5 µm. (G) Cells were sequentially infected with NUPR1 shRNA and treated with 10 nM BafA1. The graph shows the quantification of the LC3B-II:ACTB and SQSTM1:ACTB ratios in the lower panel from 3 different immunoblots (mean ± SEM). (H) Cellular morphology after sequential knockdown of NUPR1 and/or reintroduction of NUPR1 in A549 cells. Reintroduction of NUPR1 after NUPR1 depletion did not rescue the autolysosomal vacuolization phenotype. The right bar graph shows the number of vacuoles per cell. Error bars represent the SD (n = 10). N.S., not significant. Scale bars: 10 µm. (I) Immunoblot analysis of LC3B and SQSTM1 levels in A549 cells as indicated. The right bar graph depicts the densitometric analysis of protein intensity. N.S., not significant.
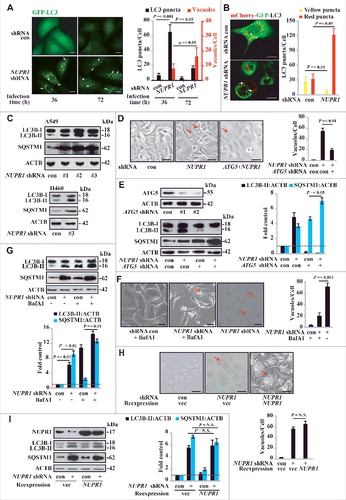
Figure 3. NUPR1 depletion causes premature senescence in vitro and represses tumorigenesis in vivo. (A) Representative images of GLB1 activity in NUPR1-depleted and control cells as indicated (left panels). Quantification of GLB1-positive cells was determined in 10 different fields from 3 independent experiments (mean ± SEM) (right panel). Scale bars: 10 µm. (B) NUPR1-depleted A549 cells were collected for cell cycle analysis by flow cytometry. The percentage of cells in G0/G1, S, and G2/M phases from 3 independent experiments is shown (right panel, mean ± SEM). (C) Western blot analysis of the indicated proteins in A549 and H460 cells infected with NUPR1 shRNA, with ACTB as a loading control. (D) Cellular proliferation of control and NUPR1-shRNA A549 cells was assessed using a 5-bromodeoxyuridine (BrdU) assay. The data are represented as the mean ± SEM of 6 experiments. (E) Clonogenic assays performed with control and NUPR1-shRNA A549 cells. A total of 1,500 cells were seeded in 24-well plates and grown for 2 wk. The graph shows the quantification of the mean number of colonies at different time point as indicated. ** P < 0.01 compared to control. (F) Western blot analysis of CASP3, cleaved CASP3, CASP7, CASP9, and ACTB in NUPR1-depleted A549 cells. (G) Western blot analysis of CDKN1B in A549 cells by NUPR1 depletion and/or its reexpression, with ACTB as a loading control. (H) A549 cells with lentivirus-delivered NUPR1 knockdown were subcutaneously implanted into female athymic nude mice (n = 6 for each experimental condition). The tumor image (left panel) on d 24 and tumor growth curve (right panel, mean ± SEM) are shown. ** P < 0.01 compared to control.
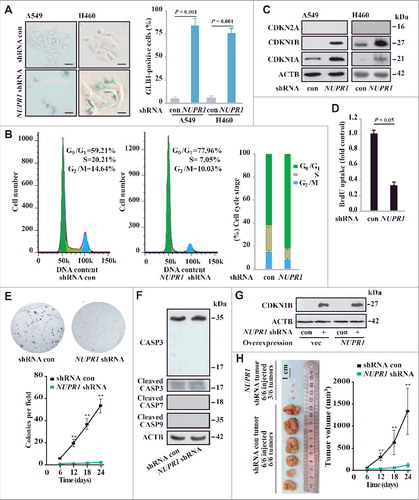
Figure 4. NUPR1 regulates autolysosomal processes through SNAP25. (A) Functional profiling of genes differentially expressed in control A549 cells and NUPR1-knockdown A549 cells. Double-headed arrows indicate 137 genes upregulated (red) and 173 genes downregulated (green) due to NUPR1 depletion. Representative related genes are listed vertically (left) and under each molecular pathway (right). (B) Quantitative RT-PCR was performed to confirm transcriptional changes of the indicated genes identified in the RNA-seq data. RNA levels were normalized to GAPDH and represent the relative fold change compared to the control shRNA samples. The mean ± SEM of 3 replicates is shown. (C) Representative phase-contrast micrographs of cell morphological changes from the indicated treatments in A549 cells. The top left panel shows SNAP25 knockdown efficiency, and the lower left bar graph shows the number of vacuoles per cell. Error bars represent the SD (n = 10). Scale bars: 10 µm. (D) Representative images of GLB1 staining in SNAP25-knockdown and control A549 cells (left 2 panels). Quantification of GLB1-positive cells was determined from 10 different fields, from 3 independent experiments (mean ±SEM) (right panel). Scale bars: 10 µm. (E) Immunoblot analysis of LC3B, SQSTM1, and ACTB in SNAP25-knockdown A549 cells.
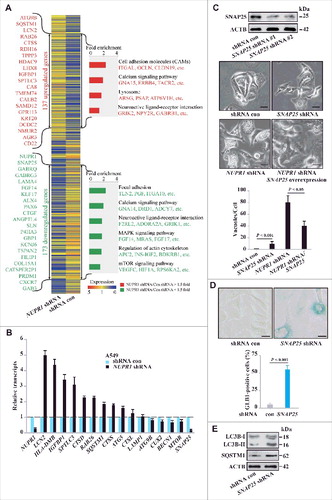
Figure 5. SNAP25 mediates autolysosomal efflux through VAMP8. (A) Immunopurification of SNAP25-containing protein complexes. Cellular extracts from A549 cells stably expressing Flag (empty vector, control) or Flag-SNAP25 were immunopurified with an M2 anti-Flag affinity gel and eluted with 3xFLAG peptide. The eluates were resolved by SDS-PAGE, and interesting bands were analyzed by mass spectrometry. *, non-specifically bound protein bands. (B) Coimmunoprecipitation results for VAMP8 and SNAP25, as well as the N- or C-terminal truncated forms, in HEK293 cells. (C) Physical interaction between SNAP25 and VAMP8 in A549 cells. Shown is the Duolink assay with the interaction between VAMP8 and STX3 as a positive control. Nuclei are counterstained with DAPI (blue). Scale bars: 10 µm. (D) Immunoblot of LC3B and SQSTM1 in SNAP25-depleted and control A549 cells, with ACTB as a loading control. (E) GLB1 staining images in VAMP8-depleted and control A549 cells. Scale bars: 10 µm. (F) Immunoblot of CTSB and CTSD in NUPR1-, SNAP25-, and VAMP8-depleted and control A549 cells, with ACTB as a loading control. (G) SNAP25 was monitored by immunoblotting after recombinant BoNT/A LC treatment at the indicated concentrations for 16 h, with ACTB as the loading control. (H) BrdU-incorporation assay in A549 cells treated with the indicated concentrations of recombinant BoNT/A LC for 24 h. (I) Representative images of GLB1 staining in BoNT/A LC-treated and control A549 cells (left panels). Quantification of GLB1-positive cells was determined from 10 different fields, from 3 independent experiments (mean ± SEM) (right panel). Scale bars: 30 µm. (J) Representative cell images of SNAP25 or VAMP8 knockdown A549 cells following 50 mM trehalose or 10 µm torin 1 treatment for 24 h. Scale bars: 10 µm. (K) Representative images of GLB1 staining in A549-knockdown cells following the indicated treatment for 24 h (left panels). Quantification of GLB1-positive cells (right panel) was determined as in (I). Scale bars: 10 µm.
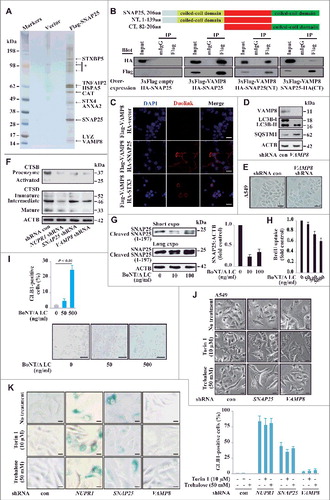
Figure 6. Histological properties of patient-derived lung NSCLC specimens. (A) Representative distribution of NUPR1 and SNAP25 determined by IHC in clinical squamous cell carcinoma and lung adenocarcinoma specimens. IHC staining with anti-NUPR1 was performed on 25 lung squamous cell carcinomas and 17 adenocarcinoma specimens compared with their adjacent tissues from the patient of origin (IHC, brown). Scale bars: 50 μm. (B) The protein level of SNAP25 was positively correlated with NUPR1 in NSCLC tissues with low (0 to 5.0 staining scores, blue lines; n = 68) versus high (5.1 to 10.0 staining scores, green lines; n = 50) NUPR1 expression (r2 = 0.332, P < 0.0001). Each red dot represents 1 tumor tissue.
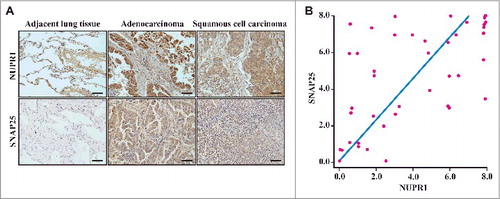
Figure 7. NUPR1 directly activates SNAP25 transcription. (A) Western blot of SNAP25, LC3B, and SQSTM1 in the indicated cell lines with ACTB as a loading control. (B) Western blot of SNAP25, BECN1/Beclin 1, MTOR, and p-MTOR (Ser2448) in A549 NUPR1-depleted cells, with ACTB as a loading control. (C) ChIP was performed to assess the association of Flag-NUPR1 with regions 4 and 5 of the SNAP25 promoter in transfected A549 cells. Stars mark ChIP primer positions. Bar graphs show fold enrichment of NUPR1 binding. Relative enrichment compared to region 1 is shown. The mean ± SEM of 3 replicates is shown. (D) The diagram shows serial deletions of the luciferase reporter upstream of the SNAP25 transcription start site (TSS); bar graphs show luciferase activity. (E) Schematic showing the balance between autophagic flux and autolysosomal efflux mediated by NUPR1 in lung cancer cells.
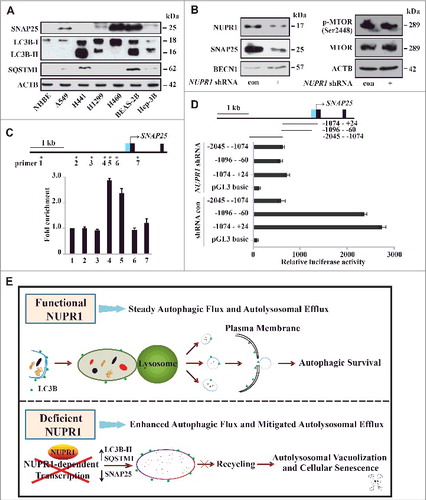
Table 1. Primary antibodies used in this study.
