Figures & data
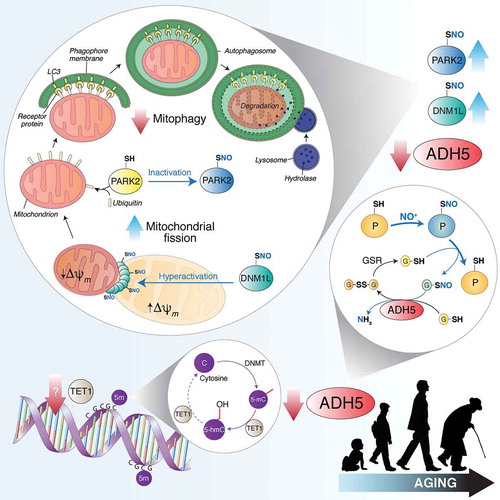
Figure 1. TET1 catalyzes the conversion of 5-methylcytosine (5-mC) into 5-hydroxymethylcytosine (5-hmC), thus counteracting gene silencing due to cytosine (C) methylation by DNMTs (DNA methyltransferases). Age-dependent TET1 decline results in ADH5 downregulation. By reducing S-nitrosoglutathione (GSNO) to glutathione disulfide (GSSG) and ammonia (NH3), ADH5 indirectly controls the amount of S-nitrosylated proteins (PSNOs), generated upon reaction with an NO moiety (in the figure shown as NO+). ADH5 decrease causes the accumulation of PSNOs, i.e., DNM1L and PARK2. DNM1L S-nitrosylation (SNO) stimulates its GTPase activity, resulting in a massive mitochondrial fragmentation. Fragmented mitochondria, characterized by low transmembrane potential (Δψm), are modified by ubiquitination on specific protein receptors by PARK2. S-nitrosylation of PARK2 inhibits its E3 ubiquitin-ligase activity, blocking, in such a way, mitochondria recognition by the autophagic machinery. Other mitophagy-related proteins (i.e., PINK1, not shown in this scheme) are sensitive to S-nitrosylation and could play a role in ADH5-dependent mitophagy control during aging.