Figures & data
Figure 1. TFEB S211 is responsible for phosphorylation-dependent interaction with YWHA/14-3-3 proteins. (a) Domain organizations of YWHA/14-3-3 proteins and TFEB. TFEB contains an N-terminal glutamine-rich domain, a transcriptional activation domain (AD) followed by a bHLH and a leucine zipper (LZ), and a C-terminal proline-rich domain. (b) Sequence alignment of human TFEB, TFE3, MITF and TFEC from the MiT/TFE family. The identical residues are colored in red and the highly conserved residues are colored in green. Notably, the extremely conserved S211 in TFEB is very close to the NLS. (c) Co-IP assay of the interactions between YWHA/14-3-3 proteins and TFEB. Both YWHAB and YWHAG can co-immunoprecipitate with wild-type TFEB. Mutations at S211 abolished the interactions but the same type of mutations at S142 did not significantly impair the binding. (d) The binding affinities between TFEB phosphorylated peptides and YWHA/14-3-3 proteins determined by ITC experiments. YWHAB and YWHAG both interact with the p-S211-peptide but not the p-S142-peptide. The binding affinities are indicated in each panel.
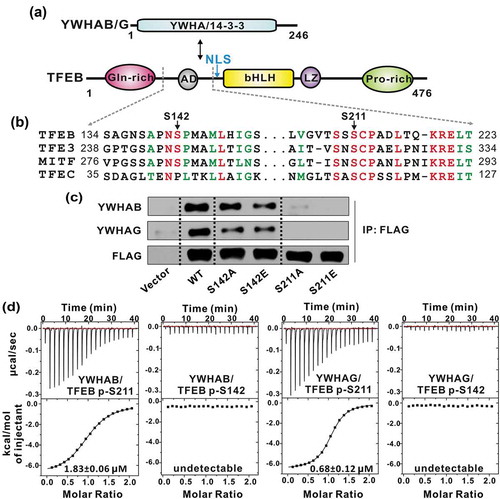
Figure 2. The overall structures of YWHA/14-3-3 proteins in complex with the TFEB p-S211-peptide. (a-b) Ribbon diagrams of the YWHAB-p-S211-peptide (a) and YWHAG-p-S211-peptide (b) complexes. YWHAB and YWHAG are colored in cyan and pink, respectively. The sidechains of the residues in the p-S211-peptide are shown as sticks. (c) Superimposition of the structures of the YWHAB-p-S211-peptide and YWHAG-p-S211-peptide complexes. (d-e) The resolved electron density maps of the p-S211-peptide in the structures of the YWHAB-p-S211-peptide (d) and YWHAG-p-S211-peptide (e) complexes. In this drawing, YWHA/14-3-3 proteins are shown as cylinders and colored as those in panel A and B. The omit electron density maps of the p-S211-peptide in the 2 structures are shown and contoured at the 1.5 σ level.
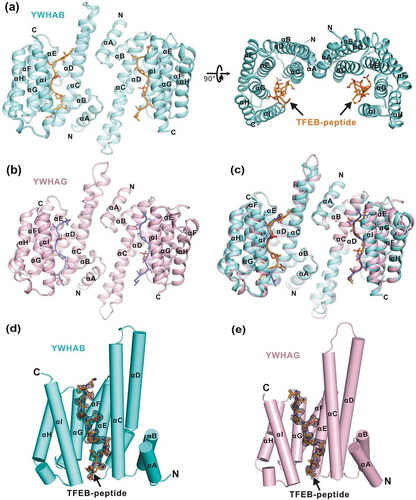
Figure 3. The interaction interface between YWHAB and the p-S211-peptide. (a) A combined surface and stick representation showing the interaction interface between YWHAB and the p-S211-peptide. The p-S211-peptide is in the stick representation (colored in orange) and YWHAB is in the surface representation. In this surface drawing, the hydrophobic, positively charged, negatively charged residues and the rest of the residues are colored in yellow, blue, red and white, respectively. The upper and lower interaction sites are highlighted by dashed boxes. (b-d) A combined ribbon-and-stick model illustrates the interaction interface between YWHAB and the p-S211-peptide. In this drawing, YWHAB and the p-S211-peptide are colored following the color patterns of and the sidechains of the residues involved in the interaction interface are shown as sticks. Hydrogen bonds and salt bridges are indicated by dashed lines.
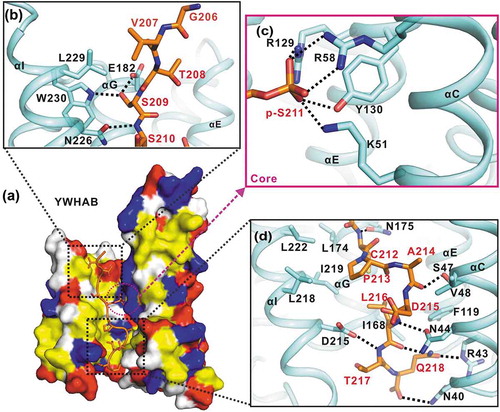
Figure 4. YWHA/14-3-3 proteins recognize TFEB p-S211-peptide by a non-canonical mode. (a-c) A combined ribbon-and-stick representation of the ‘mode I’ (PDB: 1QJB) (a), ‘mode II’ (PDB: 1QJA) (b) and ‘mode III’ (PDB: 1Q9D) (c) binding motifs for YWHA/14-3-3 proteins. The consensus binding motifs for each mode are indicated in each panel. Besides the phosphorylated-serine-mediated electrostatic interactions, R(−3) and S(−2) in mode I, R(−4) in mode II and the carboxyl tail of V(+1) in mode III also contribute to the binding. (d) TFEB binds to YWHA/14-3-3 proteins via a noncanonical mode. Compared with the canonical mode I and II binding motifs, the TFEB p-S211-peptide lacks the N-terminal arginine but contains C-terminal hydrophobic residues that bind to the lower part of the target-binding groove, which can be defined as mode IV. (e) The schematic models of the different YWHA/14-3-3-binding modes (mode I-IV). In this drawing, YWHA/14-3-3 proteins are colored in gray and their central target-binding grooves are highlighted in dark gray. The phosphorylated peptides are drawn as black lines and the key residues for binding to YWHA/14-3-3 proteins in different modes are depicted as green dots (S[−2]), blue triangles (R[−3] or R[−4]), red forks (p-S[0]), yellow pentagons (P[+2]), pink squares (V[+1]-COOH) and orange parallelogram (L[+5]). (f-g) GST affinity-isolation analysis (f) and yeast two-hybrid assay (g) of the interactions between the TFEB mutants and YWHAB. As compared to the wild-type protein, the point mutations in TFEB affected its binding to YWHAB. The interaction between TP53/p53 and T-Ag were used as the positive control for yeast two-hybrid assay. T-Ag, SV40 T antigen. (h) The binding affinities between TFEB p-S211-peptide (with mutations) and YWHAB determined by ITC experiments. The T208R mutation can enhance the binding affinity but the S209A and L216Q mutations both reduce it.
![Figure 4. YWHA/14-3-3 proteins recognize TFEB p-S211-peptide by a non-canonical mode. (a-c) A combined ribbon-and-stick representation of the ‘mode I’ (PDB: 1QJB) (a), ‘mode II’ (PDB: 1QJA) (b) and ‘mode III’ (PDB: 1Q9D) (c) binding motifs for YWHA/14-3-3 proteins. The consensus binding motifs for each mode are indicated in each panel. Besides the phosphorylated-serine-mediated electrostatic interactions, R(−3) and S(−2) in mode I, R(−4) in mode II and the carboxyl tail of V(+1) in mode III also contribute to the binding. (d) TFEB binds to YWHA/14-3-3 proteins via a noncanonical mode. Compared with the canonical mode I and II binding motifs, the TFEB p-S211-peptide lacks the N-terminal arginine but contains C-terminal hydrophobic residues that bind to the lower part of the target-binding groove, which can be defined as mode IV. (e) The schematic models of the different YWHA/14-3-3-binding modes (mode I-IV). In this drawing, YWHA/14-3-3 proteins are colored in gray and their central target-binding grooves are highlighted in dark gray. The phosphorylated peptides are drawn as black lines and the key residues for binding to YWHA/14-3-3 proteins in different modes are depicted as green dots (S[−2]), blue triangles (R[−3] or R[−4]), red forks (p-S[0]), yellow pentagons (P[+2]), pink squares (V[+1]-COOH) and orange parallelogram (L[+5]). (f-g) GST affinity-isolation analysis (f) and yeast two-hybrid assay (g) of the interactions between the TFEB mutants and YWHAB. As compared to the wild-type protein, the point mutations in TFEB affected its binding to YWHAB. The interaction between TP53/p53 and T-Ag were used as the positive control for yeast two-hybrid assay. T-Ag, SV40 T antigen. (h) The binding affinities between TFEB p-S211-peptide (with mutations) and YWHAB determined by ITC experiments. The T208R mutation can enhance the binding affinity but the S209A and L216Q mutations both reduce it.](/cms/asset/38d9d6ef-1cc7-423b-95d2-f755f040e028/kaup_a_1569928_f0004_oc.jpg)
Figure 5. The interactions between YWHA/14-3-3 proteins and TFEB are essential for TFEB subcellular localization. (a) Subcellular distribution of TFEB and its mutants under nutrient-rich and starvation conditions. Under nutrient-rich conditions, wild-type TFEB is largely distributed in the cytoplasm. The TFEB mutants (S209A, S211A, P213A, A214Q and L216Q) showed increased nuclear localization, whereas the TFEBT208R mutant did not. Upon starvation, wild-type TFEB and the TFEB mutants are mainly localized in the nucleus. Scale bar: 50 μm. (b) Quantification of the subcellular distribution data shown in panel A. The percentage of the fluorescence intensity of each construct in the nucleus was quantified (average of 4 experiments, n > 50 cells for each experiment). Each bar represents the mean ±SD, **p < 0.01, *p < 0.05. (c) A schematic model illustrating the YWHA/14-3-3-mediated regulation of TFEB subcellular localization. Briefly, the binding of YWHA/14-3-3 proteins could induce the conformational changes of the p-S211-site and the region between the NLS and the p-S211-site, which would potentially interfere with the NLS. The structural model was built based on the structures of the bHLH domain of MITF (PDB: 4ATH) and the YWHA-p-S211-peptide complex.
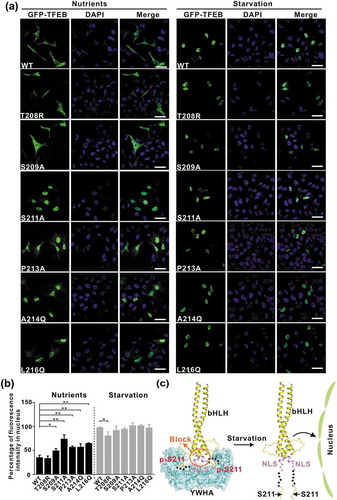
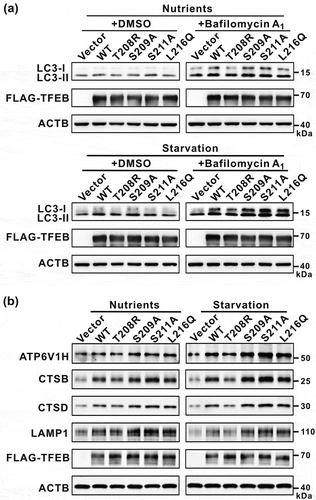
Figure 6. Modulations of the binding of TFEB to YWHA/14-3-3 proteins affect TFEB cellular functions. (a) Immunoblotting analysis of LC3 in TFEB-overexpressing cells (with wild-type TFEB and its various mutants) in the absence and presence of bafilomycin A1 under nutrient-rich and starvation conditions. (b) Western-blot analysis of the expression of TFEB target genes (ATP6V1H, CTSB, CTSD and LAMP1) after transfection with wild-type TFEB and its various mutants. ACTB was used as the loading control.