Figures & data
Figure 1. Peptide array analysis indicating LIR motifs in PIK3C3, BECN1 and ATG14. An array of 20-mer peptides covering the full-length of PIK3C3, BECN1 or ATG14, was synthesized on cellulose membrane. Each peptide was shifted 3 amino acids relative to the previous peptide. The array was probed with 1 µg/ml of GST-GABARAP for 2 h and binding was detected with anti-GST antibody. The amino acid sequence for the GABARAP interacting peptides is shown with the interacting peptides depicted as black lines. (a) Identification of two putative LIR motifs in PIK3C3 and a schematic representation of the domain organization of human PIK3C3. The locations of LIR-F198 (‘LIR’) and LIR-F250 (LIR) are indicated. (b) Identification of a minimal LIR motif in BECN1 and a schematic representation of the domain organization of human BECN1 with the LIR motif indicated. (c) Detection of two putative LIR motifs in ATG14. The domain organization of human ATG14 is schematically depicted with the locations of LIR-F64 (‘LIR’) and LIR-W435 (LIR) indicated. BH3: BCL2 homology (BH) domain 3. CC: coil-coil domain. ECD: evolutionarily conserved domain. BATS: Barkor/ATG14 autophagosome targeting sequence domain.
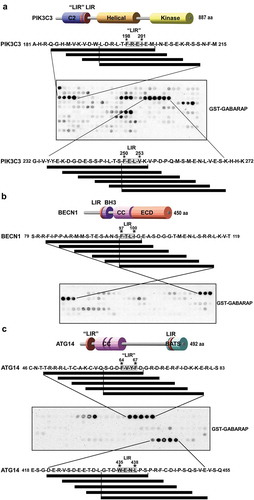
Figure 2. Verification of functional LIR motifs in PIK3C3, BECN1 and ATG14. The putative core LIR motifs, identified using peptide array analysis, were mutated by site-directed mutagenesis in the context of the full-length protein. Point mutations of both the aromatic residue and the conserved hydrophobic position were introduced in order to replace these residues with alanine. The impact of mutating the LIR motifs was then assessed in GST-pulldown analyzes with recombinant GST-Atg8 homologs and in vitro-translated 35S-labelled Myc-tagged PIK3C3, BECN1 and ATG14. Phosphomimetic mutations of the previously identified important serines (S90E, S93E and S96E) of BECN1 were made and tested for functionality with the identified LIR motif FTLI or a LIR mutant. (a) Autoradiographs (AR, top panels) and Coomassie Brilliant Blue-stained immobilized GST or GST-fusion proteins (bottom panel) are shown. (b-d) Quantifications of the binding of wild-type and mutant constructs to the GST-Atg8 proteins is presented as percentage binding relative to the 10% input. There is a significant increase in binding of the phosphomimetic mutant of BECN1 in (c). Data information: Means ± SD of 3 independent experiments. Significant P value is indicated as * = P value is 0.017 (Student´s two-tailed, unpaired t-test). ns, not significant.
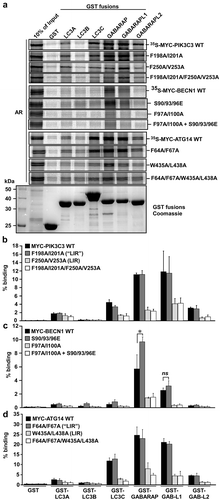
Table 1. Affinities (KD values in µM) of LIR peptides binding to ATG8 family members*.
Figure 3. Substitution analyzes of the binding of GABARAPL1 to the LIR motifs of PIK3C3, BECN1 and ATG14 using 2D peptide arrays. Two-dimensional peptide arrays scans analyzing the effect of single amino acid substitutions at all positions of the indicated 18-mer peptides containing the functional LIR motifs from (a) PIK3C3 (amino acids 242–259); (b) BECN1 (amino acids 89–106) and (c) ATG14 (amino acids 428–445). Each position of the 18-mer peptides was replaced with all 20 amino acids. The array was probed with 1 µg/ml of GST-GABARAPL1 for 2 h and binding to GST-GABARAPL1 was detected with anti-GST antibody. Asterisks indicate the location of known phosphosites. (d) Alignments of the functional LIR motifs identified in components of the PtdIns3K complex I in this work with those previously identified in the human ULK1/2 complex, Drosophila Atg1b/DmATG1B, C. elegans UNC-51 (CeUNC-51), S. cerevisiae Atg1 (ScAtg1), human ATG4B, C. elegans Lgg-3 (CeLGG-3) and S. cerevisiae Atg3 (ScAtg3). Residues at positions C-terminal to the core LIR motif that are known to interact via hydrophobic interactions with HP2 (from this study) or may do so are boxed in gray. (e) A sequence logo illustrates the distribution of amino acids at each position of the aligned sequences.
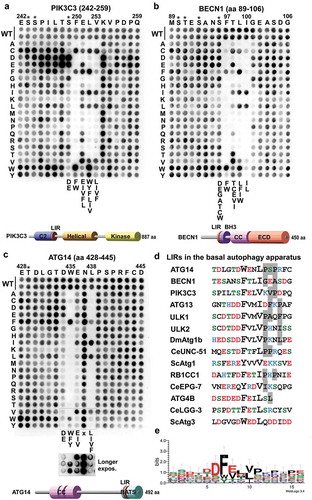
Table 2. Data collection and refinement statistics.
Figure 4. Structure of wild-type and PIK3C3S249E LIR motif bound to GABARAP. (a) Close-up of the chimera structure for the wild-type PIK3C3 LIR motif bound to GABARAP. The LIR of PIK3C3 (amino acids 244–258) is displayed in pink cartoon with the interacting residues shown as sticks. GABARAP is displayed in white cartoon and transparent surface with the hydrophobic pocket 1 and 2 colored in pink and blue surfaces, respectively. (b) Close-up of the chimera structure for PIK3C3S249E LIR motif bound to GABARAP. The LIR of PIK3C3 (amino acids 244–258) containing the phopshomimetic mutation S249E is displayed in orange cartoon with the interacting residues shown as sticks. GABARAP is displayed in white cartoon and transparent surface with the hydrophobic pocket 1 and 2 colored in pink and blue surfaces, respectively.
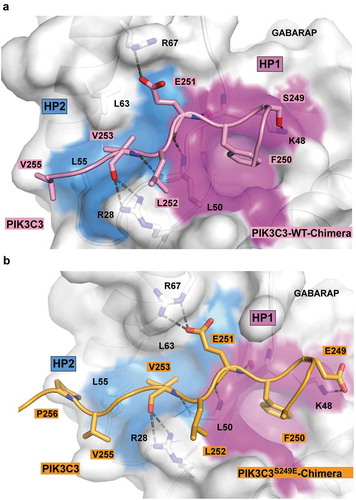
Figure 5. Structure of the BECN1 LIR motif bound to GABARAP and GABARAPL1. (a) Superposition of the chimera structure of wild type BECN1 LIR motif (yellow) bound to GABARAP and the wild type BECN1 LIR peptide (green) bound to GABARAPL1. Both LIR motifs are displayed in cartoon with the interacting residues shown as sticks. GABARAP and GABARAPL1 are displayed in white cartoon and GABARAP is displayed as well in transparent surface with the hydrophobic pocket 1 and 2 colored in pink and blue surfaces, respectively. (b) Same as (A) with the chimera structure of BECN1S96E LIR motif (cyan) bound to GABARAP superposed.
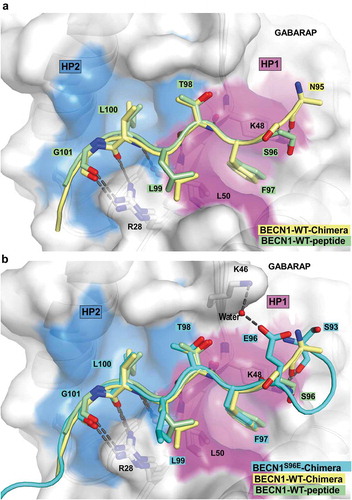
Figure 6. Structure of ATG14 LIR-W435 motif bound to GABARAPL1. (a) Close-up of the peptide structure for wild-type ATG14 LIR motif bound to GABARAPL1. The LIR of ATG14 (amino acids 429–443) is displayed in green cartoon with the interacting residues shown as sticks. GABARAPL1 is displayed in white cartoon and transparent surface with the hydrophobic pocket 1 and 2 colored in pink and blue surfaces, respectively. (b) Model of yeast complex I containing the PIK3C3 homolog Vps34, PIK3R4 homolog Vps15, BECN1 homolog Vps30 and Atg14 (PDB ID: 5DFZ) on a curved membrane [Citation6]. The membrane-proximal localization position of ATG14 and PIK3C3 LIR motifs is indicated on the figure. The structure of the part of BECN1 containing the LIR motif is not solved and therefore impossible to localize on the model.
![Figure 6. Structure of ATG14 LIR-W435 motif bound to GABARAPL1. (a) Close-up of the peptide structure for wild-type ATG14 LIR motif bound to GABARAPL1. The LIR of ATG14 (amino acids 429–443) is displayed in green cartoon with the interacting residues shown as sticks. GABARAPL1 is displayed in white cartoon and transparent surface with the hydrophobic pocket 1 and 2 colored in pink and blue surfaces, respectively. (b) Model of yeast complex I containing the PIK3C3 homolog Vps34, PIK3R4 homolog Vps15, BECN1 homolog Vps30 and Atg14 (PDB ID: 5DFZ) on a curved membrane [Citation6]. The membrane-proximal localization position of ATG14 and PIK3C3 LIR motifs is indicated on the figure. The structure of the part of BECN1 containing the LIR motif is not solved and therefore impossible to localize on the model.](/cms/asset/67dde734-d5be-4f5c-8a98-8120e49f7f3c/kaup_a_1581009_f0006_c.jpg)
Figure 7. The ATG14 LIR motif is important for binding of the PtdIns3K-C1 complex to GABARAP and GABARAPL1, and for activation of the complex by ULK1-mediated Ser29 phosphorylation of ATG14. (a) GST affinity isolation using bead-bound GST-Atg8-family fusion proteins and lysates from HCT116 cells knocked out for ATG14 and reconstituted with WT or LIR mutated MYC-ATG14. Bead bound proteins were detected by western blots using the indicated antibodies. (b) MYC-Trap experiments performed with the indicated cell lines. Bound proteins were detected by western blots using the denoted antibodies. (c) MYC-Trap experiments performed with the indicated cell lines, incubated in starvation medium (HBSS) for 6 h or not. Bound proteins were detected by western blots using the indicated antibodies. (d-g) Relative quantifications of the data in (c), showing the relative amount of BECN1, p-S93 BECN1 and p-S29 MYC-ATG14 in the MYC precipitates. Data information: Means ± SD of 3 independent experiments. Significant P values are indicated (Student´s two-tailed, unpaired t-test). ns, not significant.
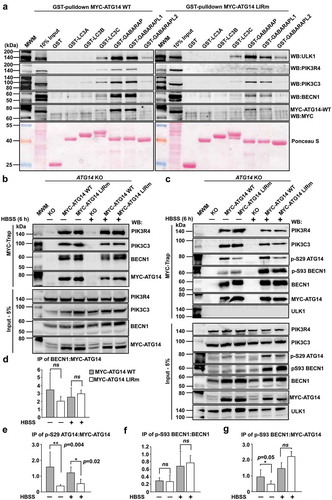
Figure 8. The ATG14 LIR interaction is important for colocalization with LC3B. (a) HCT116 cells knocked out for ATG14 and reconstituted with WT MYC-ATG14 (upper panels) or LIR mutated MYC-ATG14 (lower panels) were immunostained with MYC and LC3B antibodies and analyzed by confocal imaging. Scale bars: 10 µm and 2 µm (Enlarged). (b) Quantification of the colocalization analyzed in 8A, represented as percentage of Myc-ATG14 dots also containing LC3B. The quantification is based on analysis of 100–150 cells. (c) Quantification of EGFP intensity inside cytosolic LC3 puncta in 110–180 HeLa cells transfected with EGFP-ATG14 WT or LIR mutant. Data information: Means ± SEM; n ≥ 100 cells. Significant P values are indicated (Student´s two-tailed, unpaired t-test).
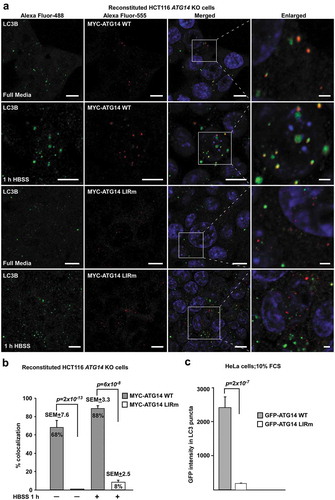
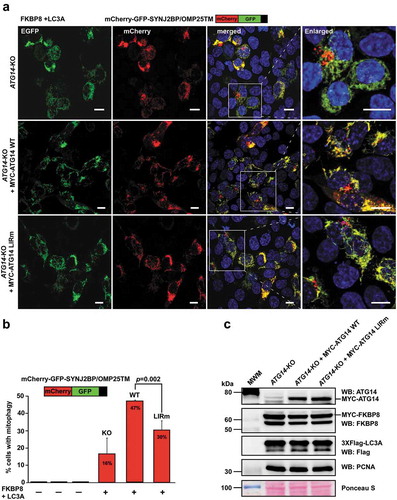
Figure 9. The ATG14 LIR interaction is important for efficient mitophagy induced by the co-expression of FKBP8 and LC3A. (a) HCT116 ATG14 KO cells reconstituted or not with WT or LIR mutated MYC-ATG14 were transiently transfected with FKBP8 and LC3A, in order to induce mitophagy, along with the mitophagy marker SYNJ2BP/OMP25-TM fused to the mCherry-GFP double tag. Mitophagy was then measured as the appearance of red-only structures. Scale bars: 10 µm. (b) Quantification of the data analyzed in 9A. The graph bars represent quantification of cells containing red‐only dots in cells expressing mCherry‐GFP‐OMP25‐TM, done manually in 60–100 cells in 3 independent experiments. Each bar shows the mean value with standard error of the mean (SEM). Two-tailed, unpaired student t-test was used to calculate p-values. (c) Western blots showing equal levels of MYC-FKBP8 and 3xFlag-LC3A expressed in the 3 cell lines. Data information: Data represent mean ± SD of 3 independent experiments (60–100 cells analyzed manually for each cell type for each experiment). Significant P values are indicated (Student´s two-tailed, unpaired t-test).