Figures & data
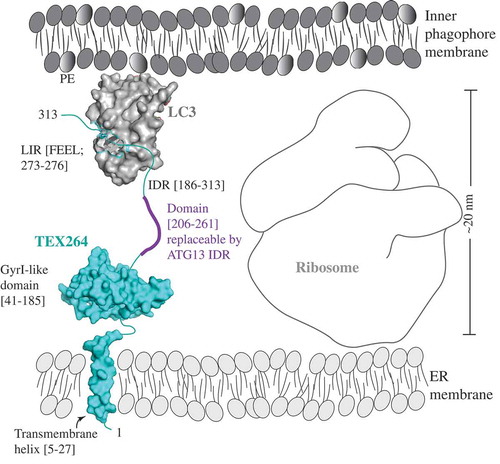
Figure 1. A model of the reticulophagy receptor TEX264 (teal) attached to the ER membrane via the transmembrane helix (residues 5–27). The gyrase inhibitor (GyrI)-like domain (residues 41–185) of TEX264 is followed by the IDR (residues 186–313) that binds via the LIR motif (F273EEL) to LC3 (gray), which is conjugated to PE at the inner phagophore membrane. The ribosome size of approximately 20 nm creates the spatial gap between the ER and inner phagophore membrane. The IDR of the small molecule TEX264 is essential for bridging this gap and for the function of TEX264 as a reticulophagy receptor. The length of the IDR linker (residues 206–261), rather than the specific amino acid sequence, is a critical factor, as shown by the finding that this segment of the TEX264 IDR is functionally replaceable by the ATG13 IDR (residues 191–248). To visualize the LIR-LC3 complex, the crystal structure of LC3 bound to the LIR of SQSTM1 was used (PDB ID: 2ZJD). The transmembrane helix and GyrI-like domain of TEX264 have been modeled using the Phyre2 server with 72% and 99.7% confidence by the single highest-scoring template c3a0hJ and d1jyha, respectively.