Figures & data
Figure 1. TXNIP is upregulated in liver tissues of NAFLD patients and MCD diet-fed mice. (A) Representative images of immunohistochemical staining for TXNIP in liver sections from controls (n = 4) and NAFLD patients (n = 11). Scale bars: 100 μm and 50 μm, respectively. A semiquantitative analysis of the IOD of TXNIP-positive areas is shown to the right of the images. Ten fields (final magnification, × 400) were randomly selected for each sample, and the IOD of all positive staining in each image was measured. (B) Association of TXNIP expression with steatosis, hepatocellular ballooning, lobular inflammation, and NAFLD activity score. (C) Representative immunohistochemical images of TXNIP protein expression in mice fed a control or MCD diet for 4 w (n = 5–7 per group). Scale bars: 100 μm and 50 μm, respectively. A semiquantitative analysis of the IOD of TXNIP-positive areas is shown to the right of the images. Ten fields (final magnification, × 400) were randomly selected for each sample, and the IOD of all positive staining in each image was measured. (D) Hepatic expression of TXNIP in mice fed a control or MCD diet (n = 5–7 per group). (E) mRNA levels of Txnip after PA treatment. AML12 cells were treated with PA (upper: 0, 200, 400, or 800 μM of PA for 6 h; lower: 400 μM PA for 0, 3, 6, or 12 h) and then subjected to qRT-PCR. (F) Subcellular localization of TXNIP after PA treatment in AML12 cells. Cells were treated with 800 μM PA for 6 h. Scale bar: 25 μm. Values represent means ± SEM. *P < 0.05, **P < 0.01 versus control
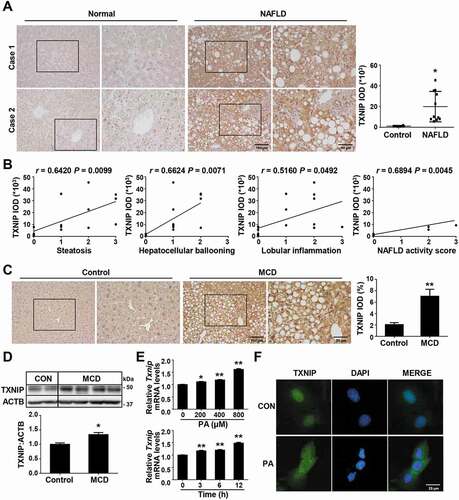
Figure 2. TXNIP deficiency accelerates MCD diet-induced liver steatosis, inflammation, and fibrosis. WT and txnip-KO mice were fed an MCD diet for 4 w (n = 5–7 per group). (A) H&E and Oil Red O staining. Scale bar: 50 μm. Ten fields (final magnification, × 400) were randomly selected for each sample, and the positive staining in each image was measured. (B) Hepatic TG levels. (C) Immunohistochemical detection of ADGRE1 (arrow). Scale bar: 50 μm. Ten fields (final magnification, × 400) were randomly selected for each sample, and positive cells in each image were counted. (D) qRT-PCR detection of mRNA levels of Adgre1, Tnf, Il6, and Il1b. (E) Sirius red staining. Scale bar: 50 μm. Ten fields (final magnification, × 400) were randomly selected for each sample, and the positive area in each image was measured. (F) Western blot analysis for ACTA2. (G) qRT-PCR detection of mRNA levels of Acta2, Tgfb, Col1a1, Timp1, and Mmp9. Values represent means ± SEM. *P < 0.05, **P < 0.01 versus WT; #P < 0.05, ##P < 0.01 versus the same-genotype control
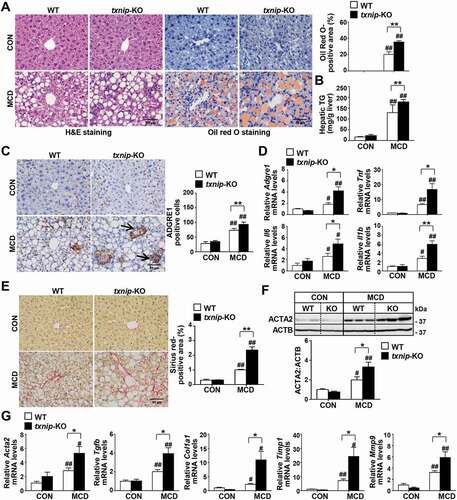
Figure 3. NAFLD patients present impaired autophagy with increased TXNIP expression. (A) Representative images of immunohistochemical staining for MAP1LC3B and SQSTM1 in liver sections from controls (n = 4) and patients with NAFLD (n = 11). Scale bars: 100 μm, 50 μm, and 5 μm, respectively. The numbers of MAP1LC3B puncta and semiquantitative analysis of the IOD of SQSTM1-positive areas are shown below the image. Ten fields (final magnification, × 400) were randomly selected for each sample, and the positive staining in each image was measured. (B) Correlation between TXNIP and MAP1LC3B or SQSTM1 expression in human NAFLD patients. Values represent means ± SEM. *P < 0.05, **P < 0.01 versus control
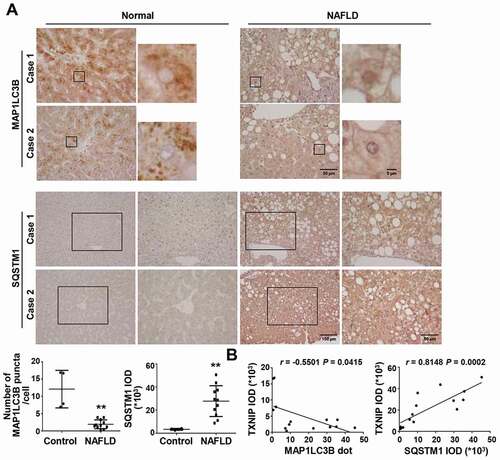
Figure 4. TXNIP deficiency causes defects in autophagy. (A) Western blot analysis of MAP1LC3B and SQSTM1 in livers of mice after 4 w of MCD diet feeding (n = 5–7 per group). (B) Immunoblot analysis of MAP1LC3B and SQSTM1 in hepatocytes after amino acid starvation. Immunoblots shown are representative of the results obtained from three independent experimental assays. (C) The formation of GFP-MAP1LC3B puncta and autophagic vacuoles. Primary hepatocytes were isolated from WT;Gfp-Map1lc3b and txnip-KO;Gfp-Map1lc3b mice. After 3 h of amino acid starvation, the numbers of puncta per hepatocyte were counted in 20 independent cells. Primary hepatocytes were also isolated from WT and txnip-KO mice. After 1 h of amino acid starvation, autophagic vacuoles were stained with green detection reagent and nuclei were stained with DAPI (blue). Scale bar: 5 μm. The numbers of autophagic vacuoles per hepatocyte were counted in 20 independent cells. (D) Autophagic vacuole formation in primary hepatocytes subjected to amino acid starvation for 1 h with or without Leu treatment. Autophagic vacuoles were stained with green detection reagent and nuclei were stained with DAPI (blue). Scale bar: 5 μm. (E) Immunoblot analysis of MAP1LC3B and SQSTM1 in hepatocytes subjected to amino acid starvation with or without Leu treatment. Immunoblots shown are representative of the results obtained from three independent experimental assays. (F) Immunoblot analysis of MAP1LC3B and SQSTM1 in AML12 control (vector control; VEC) or TXNIP-overexpressing (OE) cells. Immunoblots shown are representative of the results obtained from three independent assays. (G) Autophagic vacuole formation in AML12 cells subjected to 1 h of amino acid starvation with or without Leu treatment. Autophagic vacuoles were stained with green detection reagent and nuclei were stained with DAPI (blue). Scale bar: 15 μm. (H) Immunoblot analysis of MAP1LC3B and SQSTM1 in AML12 control (VEC) or TXNIP-overexpressing (OE) cells subjected to amino acid starvation with or without Leu treatment. Immunoblots shown are representative of the results obtained from three independent assays. Values represent means ± SEM. *P < 0.05, **P < 0.01 versus WT; #P < 0.05, ##P < 0.01 versus the same-genotype control
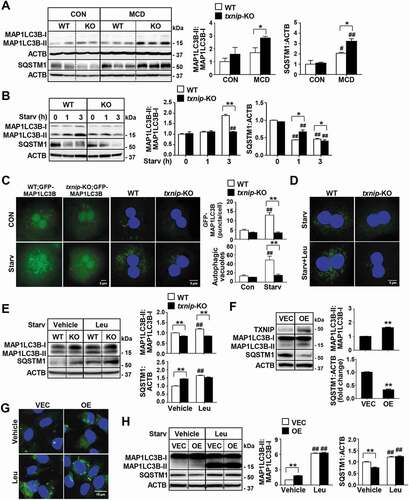
Figure 5. TXNIP deficiency causes defects in PPARA-mediated FAO. (A) Hepatic expression of FAO-related genes in mice after 4 w of MCD diet feeding (n = 5–7 per group). (B) qRT-PCR detection of mRNA levels of FAO-related genes. Primary hepatocytes were isolated and incubated with PA (125 μM) plus the PPARA agonist, WY14643 (30 μM), or DMSO for 6 h. (C) Oil Red O staining in hepatocytes. Primary hepatocytes were treated with PA (125 μM) plus WY14643 (30 μM) or DMSO for 6 h. The positive staining per hepatocyte was measured in 20 independent cells. Scale bar: 10 μm. (D) Levels of beta-hydroxybutyrate in hepatocytes. Primary WT and txnip-KO hepatocytes were treated with PA (125 μM) plus WY14643 (30 μM) or DMSO for 6 h. Values represent means ± SEM. *P < 0.05, **P < 0.01 versus WT; #P < 0.05, ##P < 0.01 versus the same-genotype control
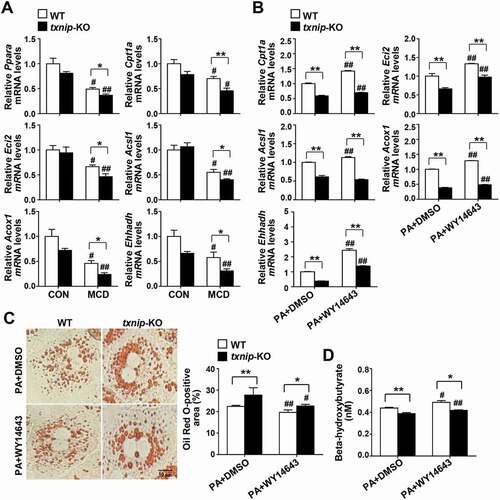
Figure 6. TXNIP induces autophagy related to the PRKAA pathway. (A) Western blot analysis of p-PRKAA and PRKAA in MCD diet-fed mice (n = 5–7 per group). (B) Immunoblotting of p-PRKAA and PRKAA in hepatocytes. Primary mouse hepatocytes from WT and txnip-KO mice were stimulated by amino acid starvation for the indicated durations. Immunoblots shown are representative of the results obtained from three independent experimental assays. (C) Western blot analysis of p-PRKAA, PRKAA, MAP1LC3B, and SQSTM1 in AML12 control (VEC) or TXNIP-overexpressing (OE) cells transfected with control (Ctrl) or Txnip siRNAs for 24 h. Immunoblots shown are representative of the results obtained from three independent assays. (D) Subcellular localization of TXNIP and p-PRKAA in AML12 cells subjected to amino acid starvation for 1 h. Scale bars: 50 μm and 10 μm. (E) Co-immunoprecipitation and immunoblot analysis of the interaction between TXNIP and p-PRKAA. HEK293 cells were transfected with Flag-TXNIP and lysates were immunoprecipitated with an anti-Flag monoclonal antibody. Co-immunoprecipitated p-PRKAA was detected with an anti-p-PRKAA antibody. (F) Western blot analysis of p-RPTOR, RPTOR, p-RPS6KB1, RPS6KB1, and nuclear TFEB in MCD diet-fed mice (n = 5–7 per group). (G) Immunoblot analysis of p-PRKAA, PRKAA, p-RPS6KB1, RPS6KB1, nuclear and cytosolic TFEB, and SQSTM1 in WT and txnip-KO hepatocytes treated with AICAR (500 µM) for 6 h and starved for 10 min. Immunoblots shown are representative of the results obtained from three independent experimental assays. Values represent means ± SEM. *P < 0.05, **P < 0.01 versus WT; #P < 0.05, ##P < 0.01 versus the same-genotype control
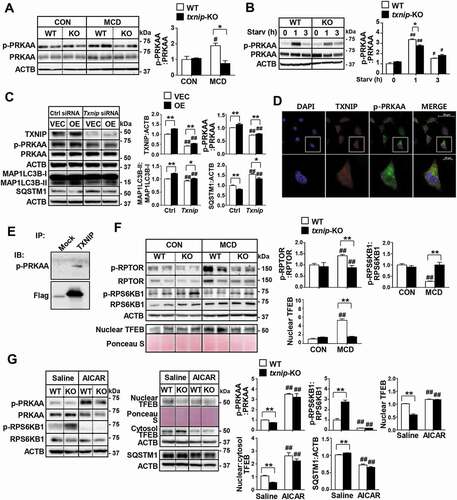
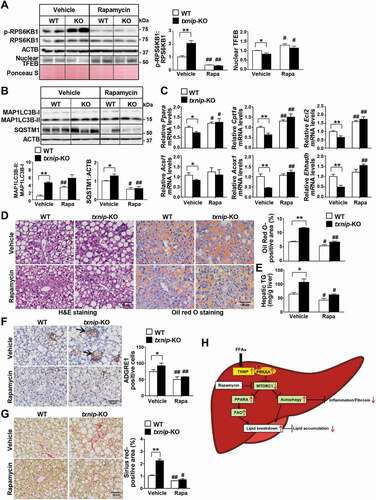
Figure 7. Induction of autophagy by MTORC1 inactivation suppresses MCD diet-induced hepatic steatosis, inflammation, and fibrosis in txnip-KO livers. WT and txnip-KO mice were fed an MCD diet for 4 w, with rapamycin (5 mg/kg/d) applied during the final week (n = 5–7 per group). (A) Western blot analysis of p-RPS6KB1, RPS6KB1, and nuclear TFEB. (B) Immunoblotting of MAP1LC3B and SQSTM1. (C) Hepatic expression of FAO-related genes. (D) H&E and Oil Red O staining. Scale bar: 50 μm. Ten fields (final magnification, × 400) were randomly selected for each sample, and the positive area in each image was measured. (E) Hepatic TG levels. (F) Immunohistochemical detection of ADGRE1 (arrow). Scale bar: 50 μm. Ten fields (final magnification, × 400) were randomly selected for each sample, and positive cells in each image were counted. (G) Sirius red staining. Ten fields (final magnification, × 400) were randomly selected for each sample, and the positive area in each image was measured. Scale bar: 50 μm. (H) A proposed model of TXNIP-mediated autophagy and FAO in NASH pathogenesis. In NASH, an elevated FFA flux upregulates TXNIP expression in hepatocytes. TXNIP promotes PRKAA phosphorylation, MTORC1 inactivation, and TFEB nuclear translocation, leading to autophagy induction and FAO. This could contribute to MCD diet-induced steatosis, inflammation, and fibrosis. Values represent means ± SEM. *P < 0.05, **P < 0.01 versus WT; #P < 0.05, ##P < 0.01 versus the same genotype control