Figures & data
Figure 1. Metabolic autophagy vs QC autophagy. (A) Metabolic autophagy is a response to diminishing sources of indicated nutrients and energy. AMPK and MTOR are at the center of responses and both along with their regulatory circuitry are located on lysosomes. Note: canonical activation of AMPK in response to AMP occurs in the cytosol (dashed arrow), whereas its noncanonical activation occurs on lysosomes in response to glucose starvation via ALDO (aldolase)-V-ATPase-AXIN-STK11/LKB1 or in response to lysosomal damage via MAP3K7/TAK1. Regulatory circuitry is indicated. TFEB is phosphorylated by MTOR and retained in the cytosol, but when nutrients are sparse and MTOR inactive, TFEB translocates to the nucleus stimulating lysosomal biogenesis. The autophagy systems (see text for definition of different protein complexes) are under control of AMPK, EP300, SIRT1 and MTOR, thought to be responsible for conducting metabolic (“bulk”) autophagy that digests macromolecules upon fusion with lysosomes to recycle/supply nutrients during starvation. Dashed box and arrow in (A) indicate engagement of components of the basal autophagy apparatus with QC autophagy in (B). (B) Quality control (QC) autophagy removes a variety of specific cytoplasmic targets, with individual cargos depicted along with their “selective autophagy” names as well as the cargo receptors implicated in each case. Both metabolic and QC autophagy contribute to cellular health and fitness.
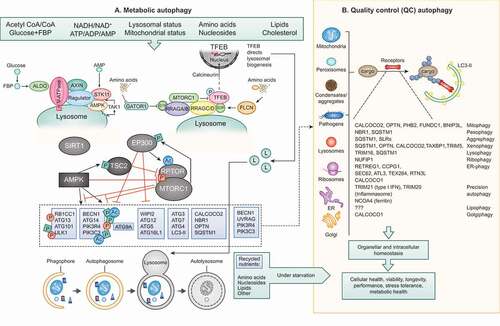
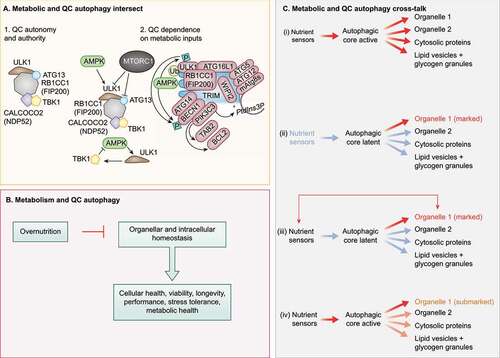
Figure 2. Crosstalk between QC and metabolic autophagy. (A) Subpanel 1. Mitophagy sponsored by CALCOCO2 can be experimentally rendered independent of upstream metabolic regulators AMPK and MTOR. Note the key contribution of the ULK1 complex, which here can be independent of its upstream regulators AMPK and MTOR, Note that CALCOCO2 remains linked to TBK1. Subpanel 2, left top. AMPK and MTOR normally control the ULK1 complex (ULK1, RB1CC1, ATG13, ATG101). Subpanel 2, left bottom. TBK1 may provide additional regulatory loops with AMPK and MTOR under physiological conditions. Subpanel 2, right. TRIMs are receptor-regulators of precision autophagy (a highly focused selective autophagy) remaining connected to the metabolic regulator AMPK. (B) Overnutrition inhibits metabolic autophagy and may compromise QC autophagy. (C) Proposed combinatorial relationships between metabolic and QC autophagy: (I) metabolic autophagy is activated by nutrient sensors acting on the autophagic core machinery and is nonselective; (ii) QC autophagy only concerns organelles (exemplified), proteins or other structures that have been marked for destruction with appropriate tags, following the detection of deficient quality (“marked”). (iii) Nutrient sensors may affect specific organelles to contribute to their marking (“marked”). (iv) When the core machinery of autophagy is activated downstream of nutrient sensors, the primary autophagic cargo may be constituted by partially deficient organelles that bear a few marking tags that usually would not lead to their destruction.