Figures & data
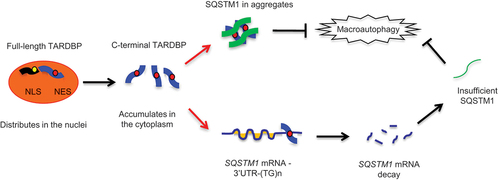
Figure 1. Cytoplasmic mutant TARDBP reduced SQSTM1 expression in the monkey brain. (A) A schematic diagram for the injection of adeno-associated viruses, which express mutant TARDBPM337V or control GFP under the CMV promoter, into the substantia nigra or motor cortex of monkeys and mouse brain. (B) Immunofluorescent staining of the monkey substantia nigra injected with AAV- GFP control (upper panel) or -TARDBPM337V (lower panel), using antibodies to GFP or FLAG (green) and SQSTM1 (red). The nuclei were stained with DAPI (blue). Representative images were obtained from three 8- to 12-years-old monkeys. The monkey brain showed significant cytoplasmic colocalization of mutant TARDBP as compared with that expressing GFP. Scale bars: 40 μm. (C) Representative immunofluorescent images of the substantia nigra in 6- to 9-months-old mice injected with AAV-TARDBPM337V or -GFP control, using antibodies to TARDBP or GFP (green) and SQSTM1 (red). The nuclei were stained with DAPI (blue). Scale bars: 40 μm. (D) Quantitative analysis of the relative numbers of cells containing cytoplasmic TARDBP over total DAPI-staining injected with AAV-TARDBPM337V or -GFP control. Twenty random fields (40X) in each section were examined from 3 samples in each group. Data are mean ±SEM. (**P < 0.01, ns: not significant). (E) Western blotting analysis of the soluble fractions in AAV-TARDBP or -GFP injected monkey and mouse tissues. Probing with antibodies to the autophagy related proteins BECN1, SQSTM1 and LC3 revealed that mutant TARDBP decreased endogenous SQSTM1 expression in the monkey brain as compared with the mouse brain. (F) The ratios of SQSTM1 to VCL, LC3-II to LC3-I, and truncated TARDBP (25-kDa) to full-length TARDBP (43-kDa) on Western blots in (E) are presented. The data were obtained from independent Western blotting analyses of tissues from AAV-GFP- or AAV-TARDBP-injected monkeys and mice (n = 3 animals per group). **P < 0.01; ns: not significant. (G) Quantitative PCR of expression of the autophagy related genes SQSTM1, LC3 and BECN1 in AAV-TARDBP monkey and mouse substantia nigra. AAV-GFP injection served as a control. Note that SQSTM1-encoding mRNA level was selectively reduced by mutant TARDBP in the monkey brain, but not in the mouse brain. The data (mean ± SE) were obtained from three independent experiments (n = 3 animals per group, *P < 0.05; **P < 0.01; ns: not significant).
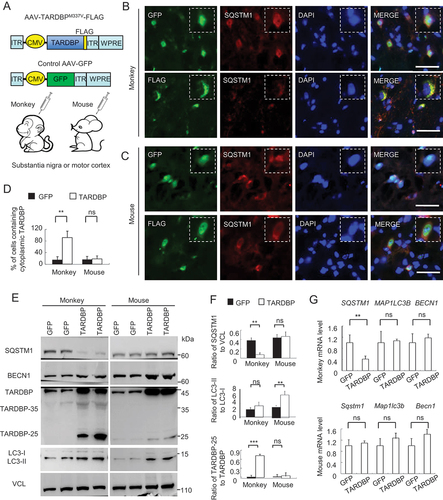
Figure 2. Preferential binding of TARDBP to the primate SQSTM1 transcript 3ʹUTR to suppress SQSTM1 expression. (A) The putative RNA binding conserved sequences “GUGUGUGUGUGU” or “GUGAGGGUGUGU” of TARDBP were determined in the box. In the 3ʹUTR region of the human, macaque and mouse SQSTM1 gene, each (GU/UG)n motif and polyA tail at the downstream of message RNA stop code were predicated. The numbers of GU/UG repeats were indicated in the boxes under the sequences. The consensus motifs with the species were indicated in red. (B) The plasmid DNA structures for expressing full-length TARDBPM337V, truncated C-terminal-TARDBP-(90-414) as TARDBP-35M337V, C-terminal-TARDBP-(184-414) as TARDBP-25M337V; or the nuclear localization signal (NLS) deletion TARDBP-ΔNLSM337V, under the same CMV promoter in pEGFP or pRK vector. (C) Ribonucleoprotein complex (RNA) immunoprecipitation (RIP) assay was performed using TARDBPM337V-FLAG transfected human SH-SY5Y and mouse Neuro-2a cells. The transfected TARDBP was immunoprecipitated by FLAG antibody and the associated mRNAs were then detected by RT-PCR. The human and mouse SQSTM1 3ʹ-UTR cDNAs were used for comparison, and their expression was seen in transfected cells and input. IgG and H2O served as control for the immunoprecipitation and RT-PCR. Compared with mouse Neuro-2a cells, human SH-SY5Y cells show that transfected TARDBP interacted with the human SQSTM1 3ʹ-UTR. (D) Western blotting analysis of the immunoprecipitated pEGFP, pEGFP-TARDBP, pEGFP-TARDBP-35, pEGFP-TARDBP-25 and pEGFP-TARDBP-ΔNLS in HEK293 cells, showed that transfected C-terminal TARDBP fragments could be precipitated by the GFP antibody. (E) The TARDBP-associated RNAs precipitated by anti-GFP (RIP) in SH-SY5Y cells were analyzed by RT-PCR with specific primers for identifying human SQSTM1 3ʹUTR (513 bp) segment. The input or IgG groups served as control. The H2O group was the negative control for the RT-PCR process. The results revealed that all the truncated cytoplasmic TARDBP, but not the GFP control, could interact with human SQSTM1 mRNA 3ʹUTR. (F) TARDBPM337V-associated RNAs were analyzed by deep sequencing (RIP-Seq) to identify the putative pre-mRNA targets. The overall distribution (left panel) and the statistic annotation (right panel) of TARDBPM337V-associated RNAs in RIP-Seq data show the enriched RNA components consisting of the CDS segment (46.65%) and the 3ʹ-UTR segment (28.91%). (G) The UCSC genome browser was used to analyze the general transcript reads of TARDBP associated RNAs. The 3ʹ-UTR segment on the human SQSTM1 gene, localized on the chromosome 5: (179,806,353 … 179,838,078), was successfully captured and amplified by deep sequencing.
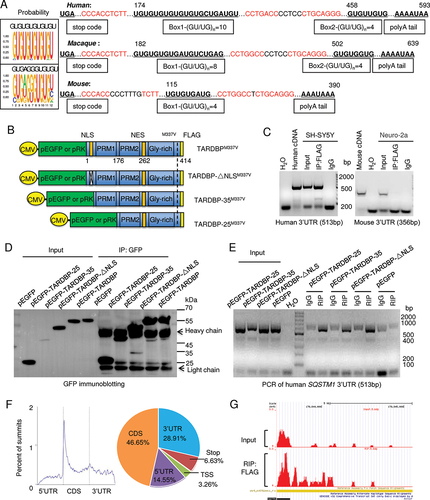
Figure 3. TARDBP-25 promoted human SQSTM1 mRNA instability and was eliminated by autophagic process. (A) The human and mouse SQSTM1 3ʹUTR segments were transfected into HEK293 cells for detection of Renilla luciferase activity. The values of luciferase report assay obtained from three independent experiments indicate that only the human SQSTM1 3ʹUTR was significantly down-regulated by mutant TARDBP, compared with the empty vector control. Data are mean ±SEM (**P < 0.01. ns: not significant). (B) The (GU/UG)n-box deletion vectors of human SQSTM1 3ʹUTR segment were generated by Site-Directed Mutagenesis Kit, then co-transfected with pRK-TARDBP-25 or empty pRK vector in HEK293 cell. The result showed that after deleting the first “(GU/UG)n = 10”, the cytoplasmic TARDBP-25 did not decrease the activity of the human SQSTM1 3ʹ-UTR when compared with the truncation of “(GU/UG)n = 4” and wild type human SQSTM1 3ʹ-UTR treatment. The values of luciferase activity were obtained from three independent experiments. Data are mean ±SEM (***P < 0.001. ns: not significant). (C) The (GU/UG)n-box (4 repeats) in the mouse Sqstm1 3ʹUTR psiCheck vector was replaced by (GU/UG)n = 10 repeats) and then co-transfected with pRK-TARDBP-25M337V into HEK293 cells. The Renilla activity of the extended mouse Sqstm1 3ʹUTR containing (GU/UG)n = 10, but not the wild type “(GU/UG)n = 4, was inhibited by cytoplasmic TARDBP-25. The values of luciferase activity were obtained from three independent experiments. Data are mean ±SE (***P < 0.001. ns: not significant). (D) The actinomycin D (ActD) chase experiment to determine endogenous mRNA’s half-lives in vitro. The human SH-SY5Y cells were transfected with pRK-TARDBP-25M337V or pRK control for 24 h and then treated for 8 h with 10 µg/ml ActD for blocking transcription. Total RNA was extracted and quantified by qPCR with specific primers for human SQSTM1. The RNA levels at different times of transfection by pPK-TARDBP-25 or pRK control were obtained by comparing that at 0 h. TARDBP-25 decreased the SQSTM1 mRNA level via its influence on mRNA stability. (E) Human SH-SY5Y cells were transfected with TARDBP siRNA or its scramble siRNA control for 48 h. Ten µg/ml ActD was added up to 8 h. The chase experiment showed that the human SQSTM1 mRNA level or its instability was not influenced by knockdown of endogenous wild type TARDBP. (F) Western blotting analysis of transfected pRK-TARDBP-25, pRK-TARDBP-35 and pRK-TARDBP in HEK293 cells after treatments with autophagic activators (rapamicin [Rapa] and LiCl) or inhibitors (bafilomycin A1 [Bafilo] and 3-MA). The blots were probed with C-terminal TARDBP antibody, revealing that TARDBP-25, but not TARDBP-35 and full-length TARDBP, was markedly reduced by autophagic activators and accumulated by the inhibitors. (G) Quantitative representation of the band intensity ratios of TARDBP-25, TARDBP-35, or TARDBP to GAPDH in (F). The data (mean ±SE) were obtained from three independent experiments. (*P < 0.05; **P < 0.01; ***P < 0.001, ns: not significant). (H) Representative staining of transfected pEGFP-TARDBP-25M337V in HEK293 cells treated with autophagic activators (rapamycin [Rapa] and LiCl) or inhibitors (bafilomycin A1 [Bafilo] and 3-MA). The immunofluorescent (upper) and phase (lower) images of the control (DMSO-treated) cells are presented. Scale bars: 20 μm. (I) Quantitative analysis of the numbers of cells in (C) containing pEGFP-TARDBP-25 per image (40X). Twenty random images in each section were examined from 3 experiments. The data are mean ±SE (*P < 0.05; **P < 0.01; ***P < 0.001, ns: not significant).
![Figure 3. TARDBP-25 promoted human SQSTM1 mRNA instability and was eliminated by autophagic process. (A) The human and mouse SQSTM1 3ʹUTR segments were transfected into HEK293 cells for detection of Renilla luciferase activity. The values of luciferase report assay obtained from three independent experiments indicate that only the human SQSTM1 3ʹUTR was significantly down-regulated by mutant TARDBP, compared with the empty vector control. Data are mean ±SEM (**P < 0.01. ns: not significant). (B) The (GU/UG)n-box deletion vectors of human SQSTM1 3ʹUTR segment were generated by Site-Directed Mutagenesis Kit, then co-transfected with pRK-TARDBP-25 or empty pRK vector in HEK293 cell. The result showed that after deleting the first “(GU/UG)n = 10”, the cytoplasmic TARDBP-25 did not decrease the activity of the human SQSTM1 3ʹ-UTR when compared with the truncation of “(GU/UG)n = 4” and wild type human SQSTM1 3ʹ-UTR treatment. The values of luciferase activity were obtained from three independent experiments. Data are mean ±SEM (***P < 0.001. ns: not significant). (C) The (GU/UG)n-box (4 repeats) in the mouse Sqstm1 3ʹUTR psiCheck vector was replaced by (GU/UG)n = 10 repeats) and then co-transfected with pRK-TARDBP-25M337V into HEK293 cells. The Renilla activity of the extended mouse Sqstm1 3ʹUTR containing (GU/UG)n = 10, but not the wild type “(GU/UG)n = 4, was inhibited by cytoplasmic TARDBP-25. The values of luciferase activity were obtained from three independent experiments. Data are mean ±SE (***P < 0.001. ns: not significant). (D) The actinomycin D (ActD) chase experiment to determine endogenous mRNA’s half-lives in vitro. The human SH-SY5Y cells were transfected with pRK-TARDBP-25M337V or pRK control for 24 h and then treated for 8 h with 10 µg/ml ActD for blocking transcription. Total RNA was extracted and quantified by qPCR with specific primers for human SQSTM1. The RNA levels at different times of transfection by pPK-TARDBP-25 or pRK control were obtained by comparing that at 0 h. TARDBP-25 decreased the SQSTM1 mRNA level via its influence on mRNA stability. (E) Human SH-SY5Y cells were transfected with TARDBP siRNA or its scramble siRNA control for 48 h. Ten µg/ml ActD was added up to 8 h. The chase experiment showed that the human SQSTM1 mRNA level or its instability was not influenced by knockdown of endogenous wild type TARDBP. (F) Western blotting analysis of transfected pRK-TARDBP-25, pRK-TARDBP-35 and pRK-TARDBP in HEK293 cells after treatments with autophagic activators (rapamicin [Rapa] and LiCl) or inhibitors (bafilomycin A1 [Bafilo] and 3-MA). The blots were probed with C-terminal TARDBP antibody, revealing that TARDBP-25, but not TARDBP-35 and full-length TARDBP, was markedly reduced by autophagic activators and accumulated by the inhibitors. (G) Quantitative representation of the band intensity ratios of TARDBP-25, TARDBP-35, or TARDBP to GAPDH in (F). The data (mean ±SE) were obtained from three independent experiments. (*P < 0.05; **P < 0.01; ***P < 0.001, ns: not significant). (H) Representative staining of transfected pEGFP-TARDBP-25M337V in HEK293 cells treated with autophagic activators (rapamycin [Rapa] and LiCl) or inhibitors (bafilomycin A1 [Bafilo] and 3-MA). The immunofluorescent (upper) and phase (lower) images of the control (DMSO-treated) cells are presented. Scale bars: 20 μm. (I) Quantitative analysis of the numbers of cells in (C) containing pEGFP-TARDBP-25 per image (40X). Twenty random images in each section were examined from 3 experiments. The data are mean ±SE (*P < 0.05; **P < 0.01; ***P < 0.001, ns: not significant).](/cms/asset/ba37cf2d-420d-4baa-83d0-cd5581f19d60/kaup_a_2013653_f0003_c.jpg)
Figure 4. SQSTM1 overexpression inhibits TARDBP-25 accumulation in vitro and in mouse brain. (A) Schematic diagrams of SQSTM1-expressing plasmid and AAV vector. The truncated AAV-TARDBP-25M337V tagged with FLAG was also expressed using AAV vector. (B) Western blotting analysis of different TARDBP fragments co-transfected with the human SQSTM1 siRNA (siSQSTM1) or control siRNA (siNC) in human SH-SY5Y cells. The knockdown of SQSTM1 increased TARDBP-25 whereas SQSTM1 overexpression decreased TARDBP-25 with no effect on TARDBP-35 or TARDBP. (C) Quantitative analysis of the band intensity ratios of TARDBP-25, TARDBP-35 and TARDBP to ACTB in (B). The data (mean ± SE) were obtained from three independent experiments (**P < 0.01; ns: not significant). (D) Immunofluorescent staining with anti-FLAG (Red) to detect TARDBP-25-FLAG and anti-SQSTM1 (green) indicated that the cytoplasmic TARDBP-25 was diminished by SQSTM1 overexpression. The nuclei were stained with DAPI (blue). Scale bar: 40 μm. (E) Quantitative analysis of the relative numbers of cells containing TARDBP-25 positive aggregates over total DAPI-nuclei per image in the injected brain region. Twenty random fields (40X) in each section were examined. n = 3 samples in each group. Data are mean ±SE. (**P < 0.01). (F) Western blotting analysis of the mouse brains injected with AAV TARDBP-25-FLAG at 6–9 months of age. The exogenous TARDBP-25-FLAG, but not endogenous TARDBP, was reduced by AAV-SQSTM1’s overexpression, as compared with the TARDBP-25-FLAG expression alone (PBS co-injection). (G) Quantitative representation of the band intensity ratios of the exogenous TARDBP-25-FLAG:GAPDH and LC3-II:LC3-I in (F). The data (mean ±SE) were obtained from three independent experiments. (**P < 0.01; ***P < 0.001, ns: not significant). (H) Immunohistochemical staining with antibody to C-terminal TARDBP showed that TARDBP-25-FLAG was also reduced by SQSTM1 overexpression. Scale bar: 20 μm. (I) Quantitative analysis of the numbers of cytoplasmic TARDBP-25-positive cells in (H). Twenty random fields (20X) in each section were examined from 3 samples in each group. Data are mean ±SEM. (**P < 0.01).
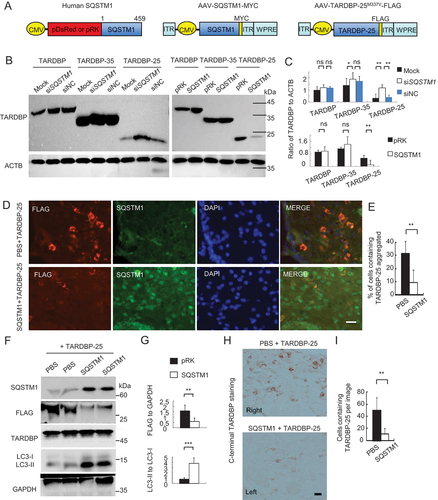
Figure 5. Overexpressing SQSTM1 promoted the clearance of TARDBPM337V in the monkey brain. (A) A schematic diagram for the stereotaxic co-injection of AAV-TARDBPM337V with AAV-SQSTM1 into the left substantia nigra and motor cortex of 10-years-old monkeys. The right side was the co-injection of TARDBPM337V with GFP control. (B) Magnetic resonance imaging (MRI) analysis revealing the BRAVO T1 and CUBE T2-weighted coronal images of the injected monkeys at the age of 10 years. The boxed areas indicate the injected regions. The right side of substantia nigra with AAV-TARDBP and AAV-GFP co-injection show damages (increased density in Sag CUBE images or reduced density in Sag BRAVO images) as compared with the left side that was co-injected with AAV-SQSTM1. (C) Western blotting with anti-C-terminal TARDBP showing that SQSTM1 overexpression promoted TARDBP-25 (arrow) clearance. Transgenic SQSTM1-MYC was detected by antibodies to MYC-Tag and SQSTM1. Note that SQSTM1 expression also increased LC3 expression. (D) Quantitative analysis of the band intensity ratios of TARDBP-25, TARDBP-35, TARDBP and LC3 to VCL in (C). The data are presented as mean ±SEM and obtained from 3 independent experiments. (*P < 0.05; **P < 0.01; ***P < 0.001, ns: not significant). (E) Immunofluorescent staining with antibodies to MYC-Tag for transgenic SQSTM1-MYC and FLAG for TARDBP-FLAG showing that SQSTM1-MYC (green) co-injection decreased the cytoplasmic accumulation of mutant TARDBP (red), in the monkey substantia nigra as compared with AAV-GFP control. Scale bar: 40 μm. (F) Quantitative analysis of the relative numbers of cells containing the cytoplasmic TARDBP-FLAG-positive cells over total DAPI-nuclei per image in the monkey substantia nigra injected with AAV-SQSTM1 or AAV-GFP control (E). Twenty random fields (40X) in each section were examined from 3 samples in each group. Data are mean ±SEM. (**P < 0.01). (G) Immunohistochemical staining with antibody to C-terminal TARDBP showing that overexpressing SQSTM1 reduced mutant TARDBP in the cytoplasm of monkey substantia nigra. Scale bar: 40 μm. (H) Quantitative analysis of the numbers of cytoplasmic TARDBP-positive cells per image (40X) in (G). Twenty random images in each section were examined from 3 samples in each group. Data are mean ±SEM. (**P < 0.01).
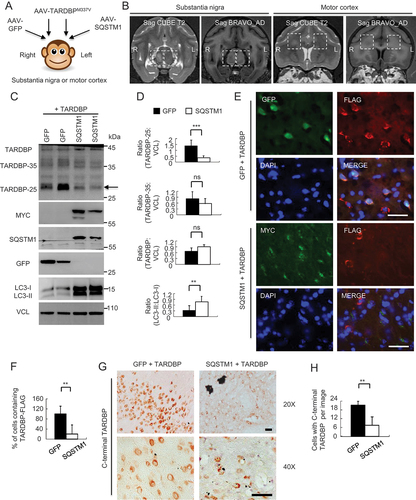
Figure 6. A proposed model for the role of SQSTM1 in clearance of cytoplasmic mutant TARDBP in the primate brain. The primate specific cleavage of TARDBP accounts for its cytoplasmic mislocalization in the primate brain. The cytoplasmic mutant TARDBP impaired the autophagic function by binding the 3ʹ-UTR of the SQSTM1 transcript to suppress SQSTM1 expression, leading to the autophagy dysfunction that can reduce TARDBP clearance in the cytoplasm. Overexpressing SQSTM1 can enhance the clearance of cytoplasmic mutant TARDBP and may be beneficial for treating TARDBP-associated neuropathology.