Figures & data
Figure 1. Atg11 is phosphorylated under autophagy induction conditions. (A) Yeast cells expressing Atg11-3×HA were grown to log phase in nutrient-rich medium (full medium), and then were treated with rapamycin for 30 min, Atg11-3×HA was detected by anti-HA antibody. (B) Yeast cells expressing Atg11-3×HA were subjected to rapamycin treatment, nitrogen starvation (SD-N), or glucose starvation (SD-G), Atg11-3×HA was detected by anti-HA antibody. (C) Yeast cells expressing Atg11-3×HA were subjected to rapamycin treatment or SD-N. Cell lysates were treated with λ protein phosphatase (λ-PPase) in the presence or absence of phosphatase inhibitors for 0.5 h at 30°C. Atg11-3×HA was detected by anti-HA antibody. Pgk1 served as a loading control.
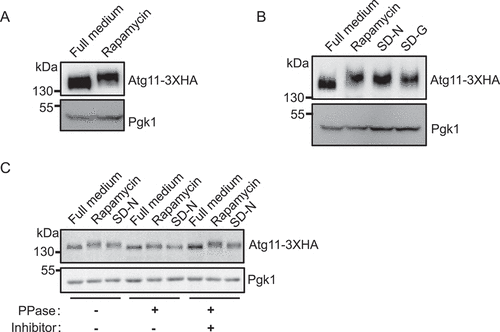
Figure 2. Atg11 is a direct phosphorylated substrate of Atg1. (A) Wild type (WT) or atg1∆ cells expressing Atg11-3×HA were grown to log phase in a nutrient-rich medium (full medium), and then were treated with or without rapamycin for 30 min. Atg11-3×HA was detected by anti-HA antibody. (B) atg1∆ yeast cells co-expressing Atg11-3×HA and empty vector, Wild-type Atg1-3×FLAG (WT), or Atg1 D211A-3×FLAG (kinase dead, KD) was subject to rapamycin treatment for 30 min. Atg11-3×HA and Atg1-3×FLAG proteins were detected by the corresponding antibody. (C) Wild type (WT), atg1∆, or atg13∆ yeast cells expressing Atg11-3×HA were subject to rapamycin treatment for 30 min. Atg11-3×HA was detected by anti-HA antibody. (D) Wild-type (WT), atg1∆, atg5∆, and atg14∆ yeast cells expressing Atg11-3×HA were subject to rapamycin treatment for 30 min. Atg11-3×HA was detected by anti-HA antibody. Pgk1 served as a loading control.
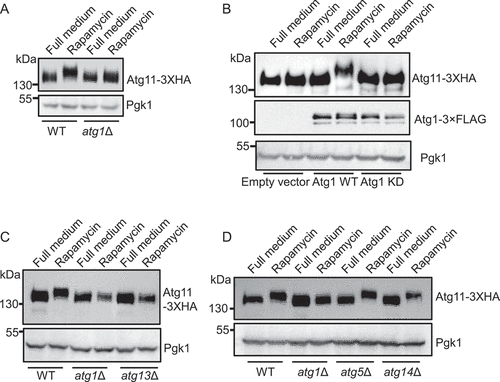
Figure 3. S949, S1057 and S1064 residues on Atg11 are phosphorylated by Atg1. (A) Secondary structure diagram of Atg11 protein. (B) GST, GST-Atg11-R1, GST-Atg11-R2, GST-Atg11-R3, and GST-Atg11-R4 were purified from E. coli via a GST column. Atg1-3×FLAG were purified from rapamycin-treated yeast cells with anti-FLAG agarose beads. in vitro kinase assays were performed with GST, GST-Atg11-R1, GST-Atg11-R2, GST-Atg11-R3, or GST-Atg11-R4 as substrates and with Atg1-3×FLAG as protein kinase. The phosphorylation level of Atg11-R1, R2, R3, or R4 was detected using anti-thioP antibody. (C) in vitro kinase assays were performed with GST -Atg11-R4 purified from E. coli as substrates and purified Atg1-3×FLAG WT or Atg1-3×FLAG KD from rapamycin-treated yeast cells as protein kinase. The phosphorylation level of Atg11 R4 was detected using anti-thioP antibody. (D) Six typical Atg1 phosphorylation sites on Ag11 R4 domain. (E) in vitro kinase assays were performed using GST, GST-Atg11-R4. The six indicated GST-Atg11 R4 variants were purified from E. coli as substrates and purified Atg1-3×FLAG from rapamycin-treated yeast cell was used as a protein kinase. The phosphorylation level of Atg11 R4 and these variants was detected using anti-thioP antibody. (F) The phosphorylation levels of Atg11 R4 were quantified and presented as mean ± SD (n = 3). ***p < 0.001; **p < 0.01; NS, no significance; two-tailed Student’s t tests were used. (G) in vitro kinase assays were performed with Atg11 R4 WT or S949A S1057A S1064A (3A) protein as substrates, which were purified from E. coli, and purified Atg1-3×FLAG from rapamycin-treated yeast cell as a protein kinase. The phosphorylation level of Atg11 R4 WT and 3A was detected using anti-thioP antibody. (H) Atg11-3×HA wild type (WT) or 3A yeast cells, and atg1∆ yeast cells expressing Atg11-3×HA were grown to log phase in nutrient-rich medium (full medium), and then were subjected to rapamycin treatment, SD-N, or SD-G for 1 h. Atg11-3×HA was detected using anti-HA antibody. Pgk1 served as a loading control.
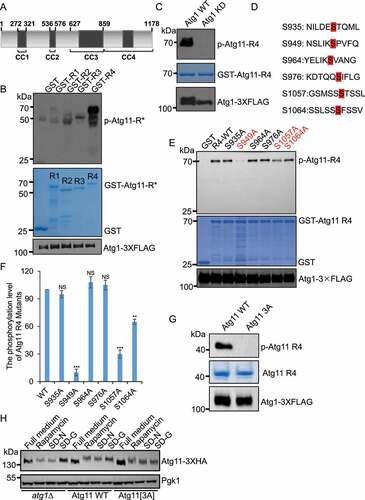
Figure 4. Atg11 phosphorylation by Atg1 is required for selective autophagy. (A) atg11∆ yeast cells, Atg11-3×HA wild type (WT) or 3A yeast cells were grown to log phase in nutrient-rich medium (full medium), and then were subjected to nitrogen starvation for 0, 2 or 4 h. The maturation of Ape1 precursor was detected using anti-Ape1 antibody. (B) Quantification of the maturation of Ape1 from (A). (C-J) atg11∆ yeast cells, Atg11-3×HA wild type (WT) or 3A yeast cells expressing Ams1 (vacuolar α-mannosidase, another the Cvt pathway substrate), Sec63 (ER protein), Pex14 (peroxisomal protein), or Om45 (mitochondrial protein) fused with GFP tag were grown to log phase, and were then subjected to nitrogen starvation for 0, 4 or 8 h. The cleavage of the indicated fusion proteins was detected using anti-GFP antibody. Pgk1 served as a loading control. The degradation rates from corresponding image were quantified and presented as mean ± SD (n = 3). ***p < 0.001; **p < 0.01; *p < 0.05; NS, no significance; two-tailed Student’s t tests were used.
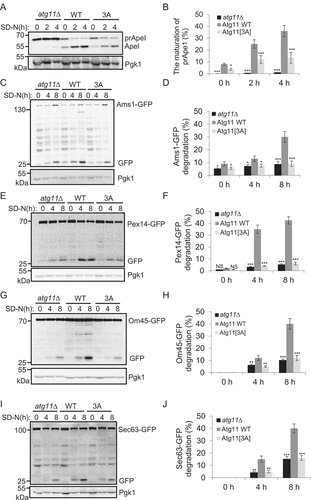
Figure 5. Atg11 phosphorylation by Atg1 regulates its association with selective receptors. (A-F) atg11∆ yeast Cells co-expressing HA-Atg11 wild type (WT) or 3A with the indicated receptor proteins fused with 3×FLAG tag was grown to log phase, then subject to SD-N for 0, 1, or 2 h. Cell lysates were immunoprecipitated with anti-FLAG agarose beads and then analyzed by western blot using an anti-HA antibody.
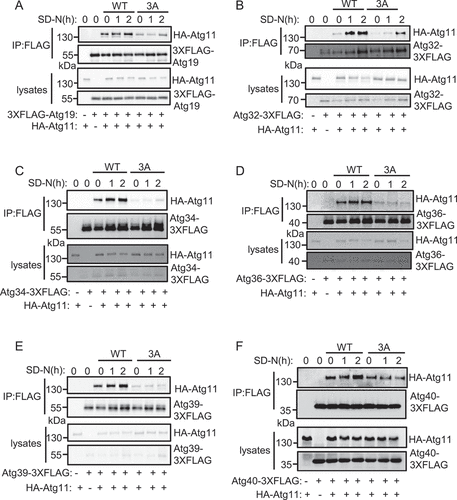
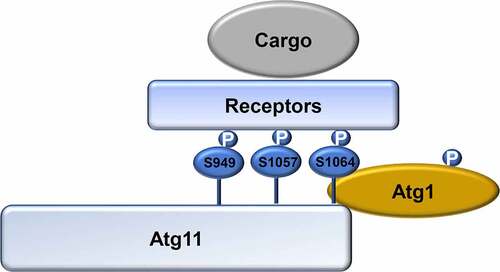
Figure 6. Model for the regulation of selective autophagy by Atg1-mediated Atg11 phosphorylation. In response to nitrogen starvation, Atg1 is activated and then phosphorylates Atg11. Phosphorylated Atg11 is required for selective autophagy by regulating the association of Atg11 with selective receptor proteins. P, phosphorylated.