Figures & data
Figure 1. G4 structures and G4 ligands. (A) Presentation of a G-quartet, a planar cyclic arrangement of four guanines held together by Hoogsteen hydrogen bonds. Stacking of two or more quartets leads to the formation of a G-quadruplex. (B) Example of a structure composed of three stacked quartets, connected by three loops – two lateral and one “chain reversal” in this example, typical of a DNA quadruplex with a so called “hybrid” topology. RNA quadruplexes tend to be all-parallel, with chain-reversal loops (for a review on quadruplex structures and topologies, please refer to [Citation106]. (C) Presentation of the G-quadruplex ligands (G4L) cited in this review; these compounds belong to chemically-distinct families. T5 is a naphtalene diimide (NDI) derivative conjugated to a carbohydrate lactose, while 3a is a naphthalimide-benzotriazole conjugate. See for details. More examples of G4L can be found in the G4L database: https://www.g4ldb.com.
![Figure 1. G4 structures and G4 ligands. (A) Presentation of a G-quartet, a planar cyclic arrangement of four guanines held together by Hoogsteen hydrogen bonds. Stacking of two or more quartets leads to the formation of a G-quadruplex. (B) Example of a structure composed of three stacked quartets, connected by three loops – two lateral and one “chain reversal” in this example, typical of a DNA quadruplex with a so called “hybrid” topology. RNA quadruplexes tend to be all-parallel, with chain-reversal loops (for a review on quadruplex structures and topologies, please refer to [Citation106]. (C) Presentation of the G-quadruplex ligands (G4L) cited in this review; these compounds belong to chemically-distinct families. T5 is a naphtalene diimide (NDI) derivative conjugated to a carbohydrate lactose, while 3a is a naphthalimide-benzotriazole conjugate. See Table 1 for details. More examples of G4L can be found in the G4L database: https://www.g4ldb.com.](/cms/asset/f8491289-08ce-4c59-8873-fe11be8383da/kaup_a_2170071_f0001_oc.jpg)
Table 1. Effect of G-quadruplex ligands on autophagy.
Figure 2. A simplified model of the lysosome-autophagy-TFEB axis. Autophagy is a lysosomal process involved in the degradation and recycling of cellular components such as proteins, lipids and mitochondria. This process is orchestrated by several proteins known as ATG (autophagy related) proteins. Autophagy occurs through multi-step processes including 1) the initiation, formation and expansion of a double-membrane structure, known as a phagophore; 2) the phagophore then encloses cytoplasmic cargoes and seals to form a closed vesicle called an autophagosome; 3) the fusion of the complete autophagosome with the lysosome to form the autolysosome; 4) the degradation of sequestered cargoes by lysosomal hydrolases. As a result, new pools of energy-rich substrates and precursors for anabolic reactions are produced. In response to several stressors (e.g., starvation, DNA damage, lysosomal stress, etc.), autophagy is activated as a result of MTORC1 inactivation. MTORC1 also coordinates the transcription activity of TFEB. Active MTORC1 recruited to the lysosome promotes the phosphorylation of TFEB that leads to TFEB sequestration in the cytosol. Conversely, when MTORC1 is inactivated, TFEB is no longer phosphorylated, allowing its translocation from the cytosol to the nucleus. As a consequence, TFEB binds to the promoter region of several lysosomal and autophagy genes containing coordinated lysosomal expression and regulation (CLEAR) sequences and promotes their transcription.
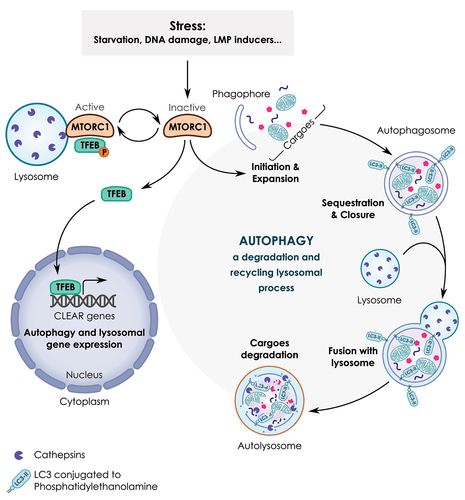
Figure 3. G4 motifs in promoters. The density of G4 motifs found in the promoters of genes involved in lysosomal function (A) or autophagy genes (B), as compared to all human promoters (unpublished data). The search for G4 motifs is made with the G4-Hunter algorithm [Citation14,Citation35]. In panel (B), “TSS-1000” and “TSS+1000” refer to the 1-kb regions upstream and downstream of the Transcription Start Site (TSS), respectively. A restricted search in the immediate vicinity of the TSS (less than 100 bp away) is also provided, as we hypothesize that the most relevant G4 are actually those within the immediate vicinity of the TSS. No statistically significant difference is found between ATG genes (blue) and all genes (green) in any of the four categories. (C) Density of G4 motifs (as deduced by G4-Hunter, shown in red) in the vicinity of the TFEB promoter (note the presence of alternative transcription start sites). An example of a quadruplex sequence with a high-Hunter score very close to one TSS is provided.
![Figure 3. G4 motifs in promoters. The density of G4 motifs found in the promoters of genes involved in lysosomal function (A) or autophagy genes (B), as compared to all human promoters (unpublished data). The search for G4 motifs is made with the G4-Hunter algorithm [Citation14,Citation35]. In panel (B), “TSS-1000” and “TSS+1000” refer to the 1-kb regions upstream and downstream of the Transcription Start Site (TSS), respectively. A restricted search in the immediate vicinity of the TSS (less than 100 bp away) is also provided, as we hypothesize that the most relevant G4 are actually those within the immediate vicinity of the TSS. No statistically significant difference is found between ATG genes (blue) and all genes (green) in any of the four categories. (C) Density of G4 motifs (as deduced by G4-Hunter, shown in red) in the vicinity of the TFEB promoter (note the presence of alternative transcription start sites). An example of a quadruplex sequence with a high-Hunter score very close to one TSS is provided.](/cms/asset/91fb9742-f0ff-465c-a416-ffb3371f1946/kaup_a_2170071_f0003_oc.jpg)
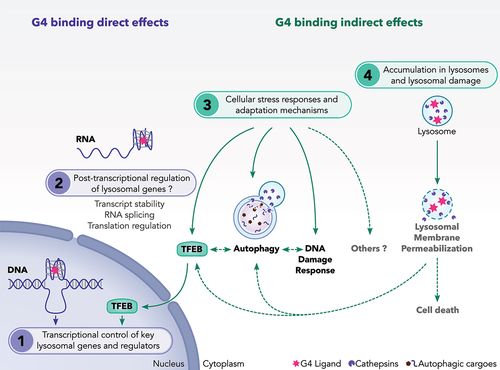
Figure 4. A proposed model showing the mechanisms of action of G4 ligands on lysosomes. We suggest that G4 ligands regulate the lysosomal pathway through multiple non exclusive mechanisms. These include: (1) Direct interaction of G4L with DNA-G4 motifs presented in the promoter of some lysosomal genes. G4L may also modulate the gene expression of TFEB transcription factor, a master regulator of lysosomal genes. (2) Direct interaction of G4L with G4 presented in RNAs relevant for autophagy and lysosomal function. So far, this regulation has not been investigated for this class of genes. (3) Activation of adaptive stress pathways such as autophagy, TFEB and DNA damage response (DDR) as a consequence of cellular stresses induced by G4L. (4) Induction of lysosomal membrane permeabilization as a consequence of the sequestration of G4L into the lysosomes. During LMP, adaptive stress responses including autophagy and TFEB are activated to cope with damaged lysosomes and to ensure cell survival. If these adaptive stress responses are unable to overcome the lysosomal stress, the LMP induction can result in cell death.