Figures & data
Figure 1. Heat map illustrating percentage of methylation for all differentially methylated cytosines (DMCs) identified (a) between genotypes, (b) between environments, (c) between environments for DAN strain, and (d) between environments for R strain (logistic regression q < 0.01 and |ΔM|>20%, and t.test p < 0.01) using unsupervised hierarchical clustering. Rows represent a unique CpG site and columns individual fish.
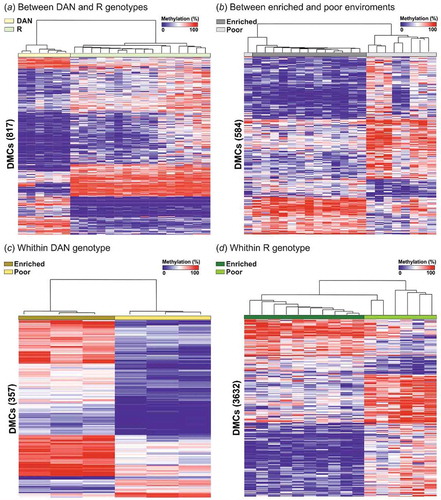
Table 1. Methylation differences averaged (percentage) for differentially methylated cytosines (DMCs) between environments (poor, enriched), shared between genotypes (DAN, R) which overlap annotated genes (reference genome ASM164957v1, GCA_00164975.1). Epiallele classification (pure or facilitated) followed [Citation9]. Positive and negative values represent increased and decreased methylation in enriched and poor environments, respectively. Q-value is the p-value adjusted by false discovery rate (FDR = 0.05).
Table 2. Linear model of principal component scores for mangrove killifish epialleles shared between genotypes (R, DAN) and environments (poor, enriched).
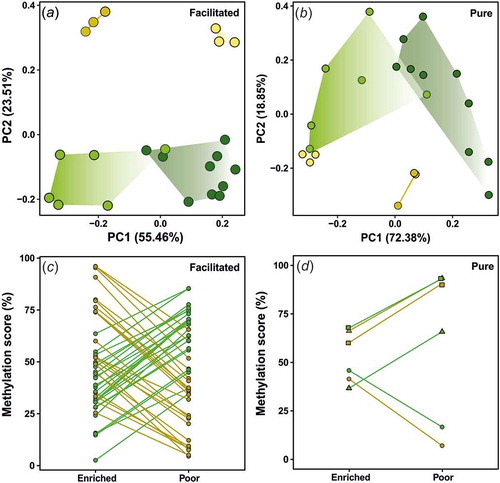
Figure 2. Principal component analysis (PCA) and reaction norms of epialleles across genotypes and environments. PCAs were based on individual scores of methylation across either (a) facilitated or (b) pure annotated DMCs. Dark yellow for DAN individuals on enriched environments; light yellow for DAN genotype on poor environments; dark green for R individuals on enriched environments; light green for R genotype on poor environments. Each reaction norm represents the change on averaged methylation scores (in percentage) for (c) facilitated and (d) pure epialelles annotated DMCs across environments. Different colours represent the genotypes (yellow for DAN; green for R). Different shapes (d) represent different annotated DMCs. Epialelles were classified according to Richards (2006). Detailed information for each annotated DMCs methylation score across genotypes is available in .