Figures & data
Figure 1. Example of phenotypic abnormalities of SLOS and ART-LOS calves. (a) Abdominal wall defect of US_SLOS_#5 (Angus breed). This spontaneous LOS calf was born alive and had to be euthanized due to the body wall malformation. (b and c) Macrosomia and macroglossia of US_SLOS_#6 (Charolais breed). This stillborn calf was ~77 Kg at birth. The average weight for calves of this breed is ~ 36 Kg. (d and e) Macroglossia of ES_ART_#2 and of stillborn ES_RF_necropsy_#1 (Asturian Valley x Limousin crossbred), respectively.
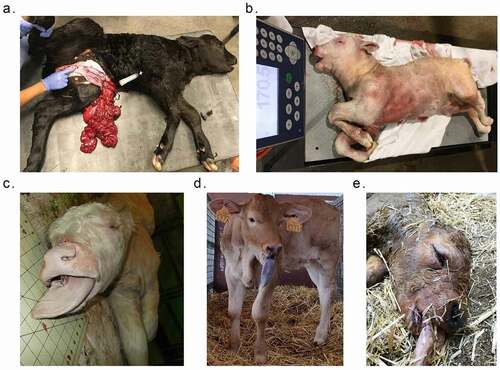
Table 1. Information of calves in this study. Sex with * = predicted sex based on WGBS reads alignment to chromosome Y (lack of sex information from original owners/sample providers). BW = birth weight. Used ID = animal ID used in previous publications.
Figure 2. Distribution of LOS associated differentially methylated regions (DMRs) across various genomic contexts. (a-c) Muscle US_SLOS vs. US_Control DMRs. (d-f) Muscle ES_RF_necropsy vs. ES_Control DMRs. (a-b and d-e) Each figure shows the total number of DMRs in the comparison and the number and percent of the hypermethylated (hyper; a and d) and hypomethylated (hypo; b and e) DMRs over each genomic context. In addition, the figures include the number and percent of DMRs that overlap with two previous studies (Li [Citation25] and Chen [Citation19]) for comparison purposes. (c and f) Percent of the genomic context that overlaps with DMRs. Obs = observed frequencies. Exp = expected frequencies (mean ± standard deviation; obtained from randomly shuffling DMRs across genome 10,000 times). The p values were calculated as p = n(|Exp – mean(Exp)| ≥ |Obs – mean(Exp)|)/10,000.
![Figure 2. Distribution of LOS associated differentially methylated regions (DMRs) across various genomic contexts. (a-c) Muscle US_SLOS vs. US_Control DMRs. (d-f) Muscle ES_RF_necropsy vs. ES_Control DMRs. (a-b and d-e) Each figure shows the total number of DMRs in the comparison and the number and percent of the hypermethylated (hyper; a and d) and hypomethylated (hypo; b and e) DMRs over each genomic context. In addition, the figures include the number and percent of DMRs that overlap with two previous studies (Li [Citation25] and Chen [Citation19]) for comparison purposes. (c and f) Percent of the genomic context that overlaps with DMRs. Obs = observed frequencies. Exp = expected frequencies (mean ± standard deviation; obtained from randomly shuffling DMRs across genome 10,000 times). The p values were calculated as p = n(|Exp – mean(Exp)| ≥ |Obs – mean(Exp)|)/10,000.](/cms/asset/d4299e45-fe8d-488e-93ce-afd412b93cc4/kepi_a_2067938_f0002_oc.jpg)
Figure 3. Example of LOS-associated vulnerable loci. Hypo = hypomethylation. Hyper = hypermethylation. Differentially methylated regions (DMR) identifiers are the positions in the bovine genome assembly ARS-UCD1.2. For complete information please refer to Table S2.
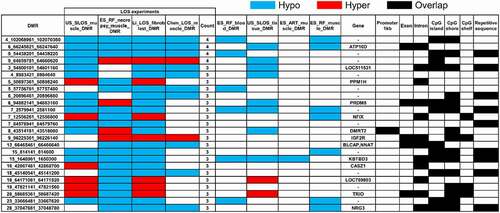
Figure 4. LOS-vulnerable loci around promoter regions. This figure shows DNA methylation level of vulnerable loci 7_2579941_2581100 (a), 8_43514181_43518080 (b), and 9_64659781_64660620 (c) in four LOS experiments. The aforementioned numbers refer to the chromosomes and genomic position in bovine genome assembly ARS-UCD1.2. Met% = group mean CpG methylation level in percent. Cov = group mean CpG read coverage. DMR = differentially methylated regions. Hyper = hypermethylation (red). Hypo = hypomethylation (yellow).
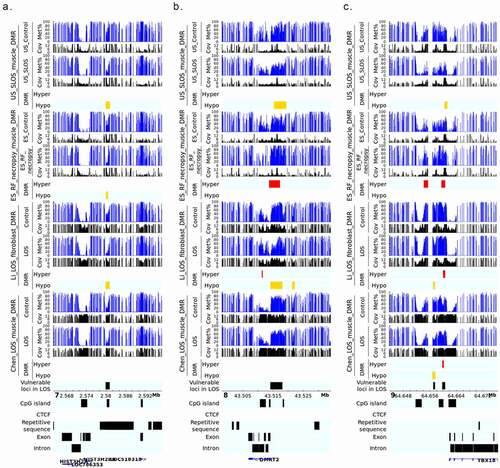
Figure 5. LOS-vulnerable loci overlapping CpG Islands in body of imprinted genes. This figure shows DNA methylation level of vulnerable loci 6_94882141_94883160 (a), 9_96225361_96226140 (b), and 13_66465461_66466640 (c) in four LOS experiments. The aforementioned numbers refer to the chromosomes and genomic position in bovine genome assembly ARS-UCD1.2. Met% = group mean CpG methylation level in percent. Cov = group mean CpG read coverage. DMR = differentially methylated regions. Hyper = hypermethylation (red). Hypo = hypomethylation (yellow).
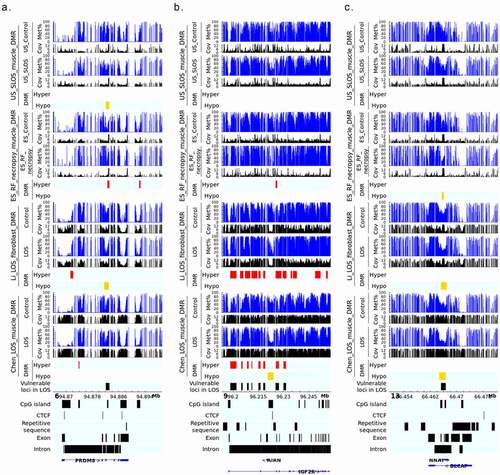
Figure 6. DNA methylation at the 25 highly vulnerable loci of LOS (≥ 3 experiments) in US_SLOS_#6 calf, its sire, dam, and full-sibling. (a) Violin plots with dots showing the difference of DNA methylation between examined individual/group (mean) and mean of US_Control for highly vulnerable loci in LOS (found in ≥3 LOS experiments). Each dot in the violin plot represents a vulnerable locus. P values were from t-test. The baseline for blood samples is US_Control blood and the baseline for muscle samples is US_Control muscle. (b-p) Box plots with dots show DNA methylation level (y-axis) at highly vulnerable loci without obvious differences (>10%) in parental blood samples (b-f), with obvious differences in sire only (g), both sire and dam (h-i), dam only (j-o), and sibling only (p) when compared to the mean of control group. Each dot in the box plot represents a sample.
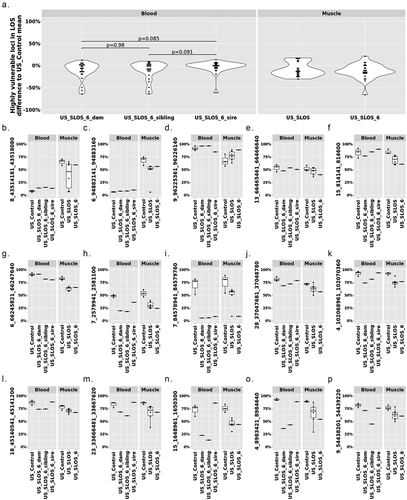
Figure 7. Distribution of ART associated differentially methylated regions (DMRs) across various genomic contents. (a-c) Muscle ES_ART vs. ES_Control DMRs. (d-f) Muscle ES_RF vs. ES_Control DMRs. (a-b and d-e) Each figure shows the total number of DMRs in the comparison and the number and percent of the hypermethylated (hyper; a and d) and hypomethylated (hypo; b and e) DMRs over each genomic context. In addition, the figures include the number and percent of DMRs that overlap with two previous studies (Li [Citation25] and Chen [Citation19]) for comparison purposes. (c and f) Percent of the genomic context that overlaps with DMRs. Obs = observed frequencies. Exp = expected frequencies (mean ± standard deviation; obtained from randomly shuffling DMRs across genome 10,000 times). The p values were calculated as p = n(|Exp – mean(Exp)| ≥ |Obs – mean(Exp)|)/10,000.
![Figure 7. Distribution of ART associated differentially methylated regions (DMRs) across various genomic contents. (a-c) Muscle ES_ART vs. ES_Control DMRs. (d-f) Muscle ES_RF vs. ES_Control DMRs. (a-b and d-e) Each figure shows the total number of DMRs in the comparison and the number and percent of the hypermethylated (hyper; a and d) and hypomethylated (hypo; b and e) DMRs over each genomic context. In addition, the figures include the number and percent of DMRs that overlap with two previous studies (Li [Citation25] and Chen [Citation19]) for comparison purposes. (c and f) Percent of the genomic context that overlaps with DMRs. Obs = observed frequencies. Exp = expected frequencies (mean ± standard deviation; obtained from randomly shuffling DMRs across genome 10,000 times). The p values were calculated as p = n(|Exp – mean(Exp)| ≥ |Obs – mean(Exp)|)/10,000.](/cms/asset/f9e8b194-76cc-4bae-ae01-8a7c274a2891/kepi_a_2067938_f0007_oc.jpg)
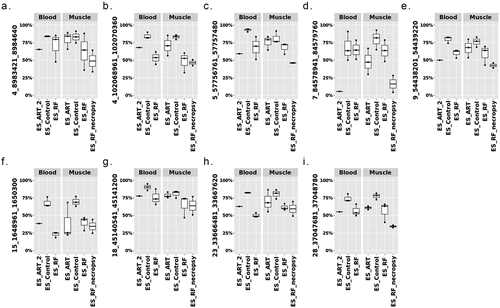
Figure 8. Conservation of DNA methylation at LOS-vulnerable loci between muscle and blood in ES_RF group. Y-axis = DNA methylation level. Note: ES_ART_2 is the same animal as ).
Supplemental Material
Download Zip (15.9 MB)S equencing d ata a vailability statement
The raw sequencing reads of WGBS used in this study are available in the GEO database with accession numbers (GSE199084).
