Figures & data
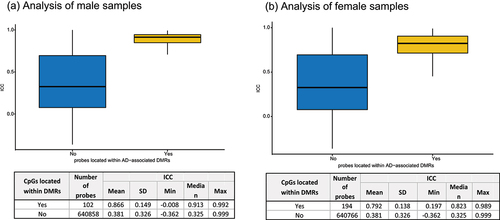
Figure 1. Distribution of estimated intraclass correlation coefficient (ICC) of DNA methylation levels in EPIC-EPIC comparison, using 69 blood DNA samples, each measured twice, generated by the Alzheimer’s disease neuroimaging initiative study. Dashed lines indicate mode of the distribution.
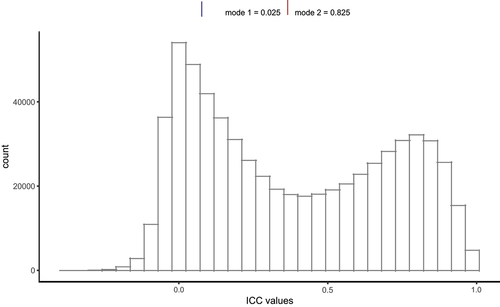
Figure 2. Probe reliability (ICC) increased as standard deviation (SD) of DNA methylation levels increased.
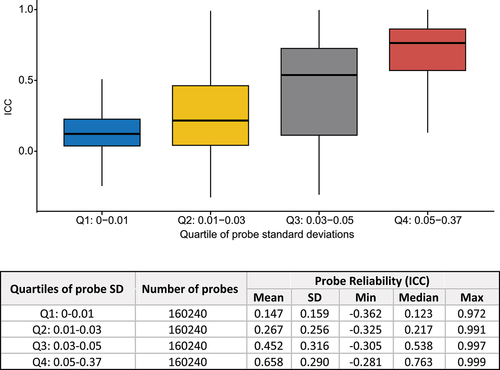
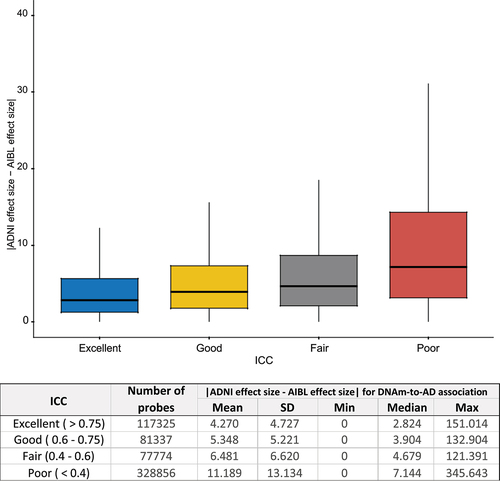
Figure 3. Higher probe reliability (ICC) is associated with smaller absolute difference in the estimated effect sizes of DNAm-to-AD diagnosis associations in ADNI and AIBL studies (p < 2.2 × 10−16). The effect sizes for DNAm-to-AD associations were obtained from Silva et al. (2022) (PMID: 35982059). Reliability of the probes were determined based on ICC: excellent (>0.75), good (0.6–0.75), fair (0.4–0.6), or poor (<0.4). AD: Alzheimer’s disease.
Supplementary Figures_1_27_2024.docx
Download MS Word (102.8 KB)Supplementary Table 6.xlsx
Download MS Excel (68.9 MB)Supplementary Tables_1_31_2024.docx
Download MS Word (50.6 KB)Data availability statement
The ADNI datasets can be accessed from http://adni.loni.usc.edu. The scripts for the analysis performed in this study are available at https://github.com/TransBioInfoLab/DNAm-reliability