Figures & data
Table 1. The sequence characteristics of the identified SABATH genes in tea plant.
Figure 1. Phylogenetic, gene structure and conserved motif analyses of CsSABATHs. (a) Phylogenetic relationships of the 32 CsSABATH proteins. (b) Exon-intron structures of CsSABATH genes. (c) The conserved motifs of CsSABATH proteins.
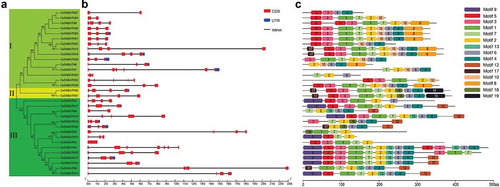
Figure 2. Phylogenetic analysis of SABATH proteins in different plants. The accession number of each protein is listed in Table S2.
Figure 3. The statistics of the major stress-related and hormonal response cis-regulatory elements detected in the promoter regions of CsSABATH genes.
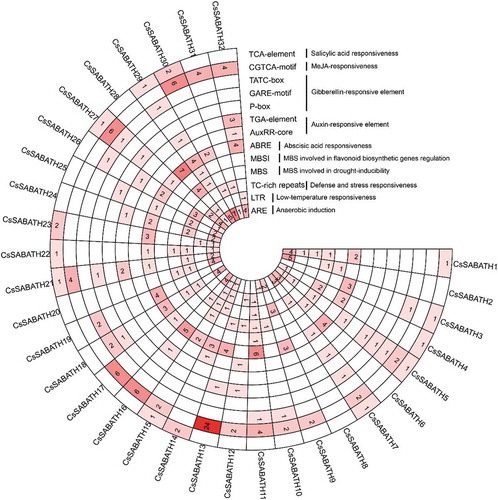
Figure 4. The spatiotemporal expression patterns of SABATH genes in tea plant. (a) The expression patterns of CsSABATHs in eight different tissues of tea plant. (b) The expression patterns of CsSABATHs in different seasons of tea leaves.
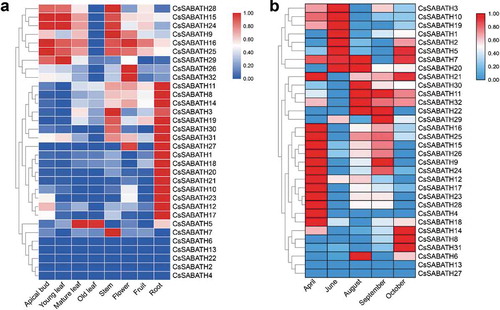
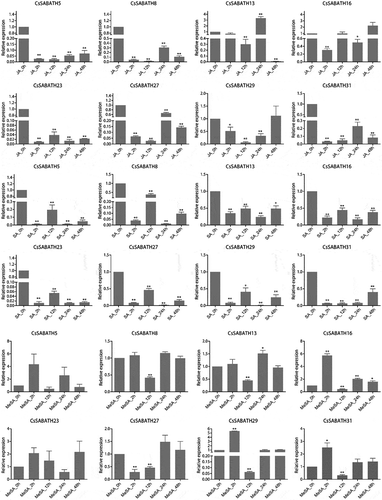
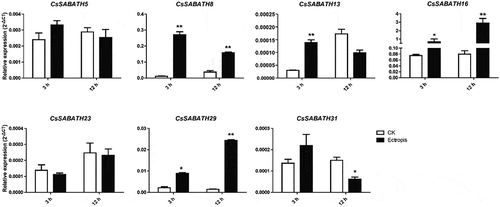
Figure 5. The stress response expression patterns of CsSABATHs in tea leaves. (a) The expression patterns of CsSABATHs in response to cold acclimation. The five stages represent nonacclimated at 25 ~ 20°C (CK), fully acclimated at 10°C for 6 h (CA1-6 h) and 10 ~ 4°C for 7 days (CA1-7d), cold response at 4 ~ 0°C for 7 days (CA2-7d) and recovering under 25 ~ 20°C for 7 days (DA-7d). (b) The expression patterns of CsSABATHs in response to 25% PEG treatment for 0, 24, 48 and 72 h. (c) The expression patterns of CsSABATHs in response to 200 mM NaCl treatment for 0, 24, 48 and 72 h. (d) The expression patterns of CsSABATHs in response to 0.25% (v/v) water solution of MeJA treatment for 0, 12, 24 and 48 h. (e) The expression patterns of CsSABATHs after Ectropis obliqua feeding in tea leaves.
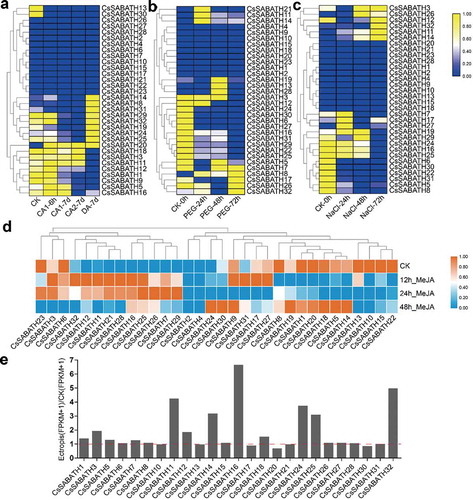
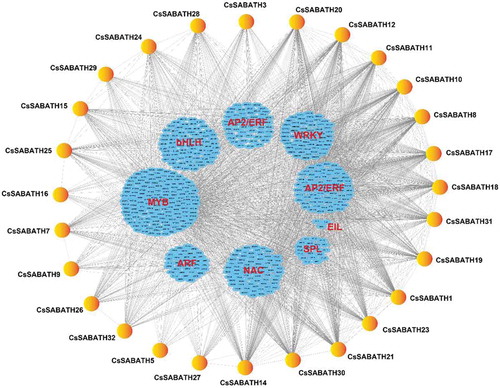
Figure 6. Transcriptional regulation of CsSABATH genes. The coexpression network of CsSABATH genes with MYB, ARF, NAC, SPL, EIL, AP2/ERF and WRKY transcription factors was constructed based on their expression patterns in eight different tissues of tea plant in Cytoscape v3.7.1. The expression correlations between CsSABATH genes and different transcription factors are shown with gray lines (Pearson’s correlation test, P ≤ 0.05), and their detailed correlations are listed in Table S3.