Figures & data
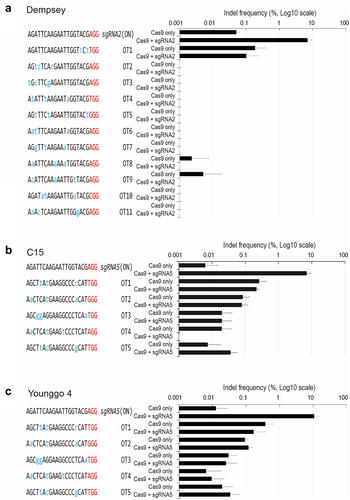
Figure 1. In vitro cleavage assay for CRISPR/Cas9 RNP-mediated CaPAD1 editing in three pepper cultivars.
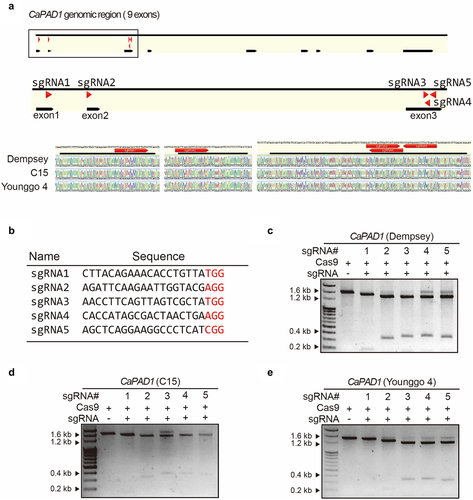
Figure 2. Three in vitro cultured pepper cultivars and RNP-delivered pepper protoplasts.
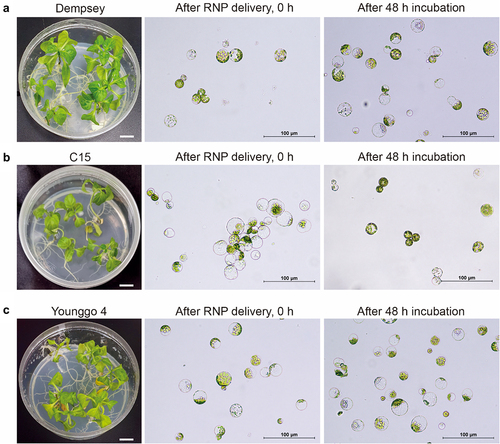
Figure 3. Analyses of CRISPR/Cas9 RNP-mediated CaPAD1 editing in three pepper cultivars.
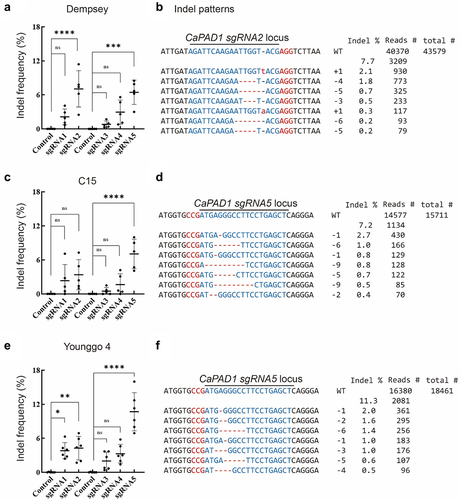
Figure 4. In vivo analyses of CaPAD1-sgRNA2 or CaPAD1-sgRNA5 activities at potential off-target sites across three pepper genomes.