Figures & data
Table 1. The primer sequences of genes.
Figure 1. Change and characterization of circ_0044907 in AML. (A) Fold change of circ_0044907 in AML patient-derived BM relative to healthy control-derived BM was determined by RT-qPCR. (B) Diagnostic value (ROC) of BM circ_0044907 in 45 AML patients. (C) Correlation between BM circ_0044907 expression pattern and clinical survival rate was analyzed. (D) Relative expression changes of circ_0044907 in AML cells. (E) Analysis of circ_0044907 and linear RPS6KB1 in AML cells treated with or without RNase R. ***P < 0.001.
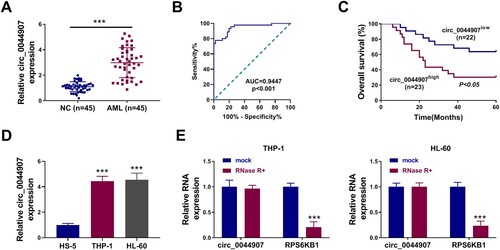
Table 2. Correlation of circ_0044907 expression with AML genotype.
Figure 2. Circ_0044907 silencing decreased AML cell proliferation and contributed to AML cell apoptosis. (A) Analysis of the interference efficiency of 3 siRNAs targeting circ_0044907 (si-circ_0044907#1, si-circ_0044907#2, and si-circ_0044907#3) was done. (B-F) MTT, EdU, and flow cytometry assays were utilized for analysis of cell viability (B), proliferation (C), apoptosis (D), and cycle progression (E and F) in AML cells with si-NC or si-circ_0044907#1. (G) Relative protein levels of PCNA, Cyclin D1, and Cleaved caspase 3 in the above AML cells. **P < 0.01 and ***P < 0.001.
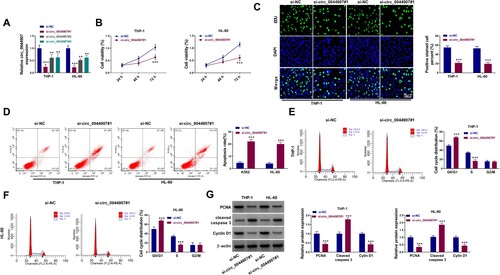
Figure 3. MiR-186-5p was sponged by circ_0044907. (A) The putative binding sites of miR-186-5p in the circ_0044907 sequence. (B) The transfection efficiency of miR-186-5p mimic. (C and D) Luciferase activity of circ_0044907-WT/circ_0044907-MUT reporter in AML cells co-transfected with miR-186-5p mimic or miR-NC. (E and F) The interaction between circ_0044907 and miR-186-5p was demonstrated by RNA pull-down and RIP assays. (G) Relative expression changes of miR-186-5p in AML patient-derived BM. (H) Pearson’s correlation coefficient analysis of the correlation between miR-186-5p and circ_0044907 in AML patient-derived BM. (I) Relative expression levels of miR-186-5p in AML cells. (J) Influence of circ_0044907 silencing on miR-186-5p in AML cells. ***P < 0.001.
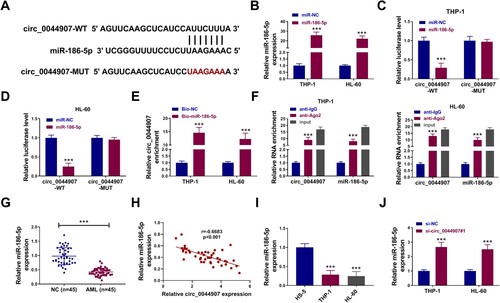
Figure 4. Circ_0044907 mediated AML cell proliferation and apoptosis through interacting with miR-186-5p. (A) The transfection efficiency of miR-186-5p inhibitor. (B-F) Analysis of cell viability (B), proliferation (C), apoptosis (D), and cycle progression (E and F) in AML cells with si-NC, si-circ_0044907#1, si-circ_0044907#1 + anti-NC, or si-circ_0044907#1 + anti-miR-186-5p. (G) Detection of PCNA, Cyclin D1, and Cleaved caspase 3 protein levels in the above AML cells. **P < 0.01 and ***P < 0.001.
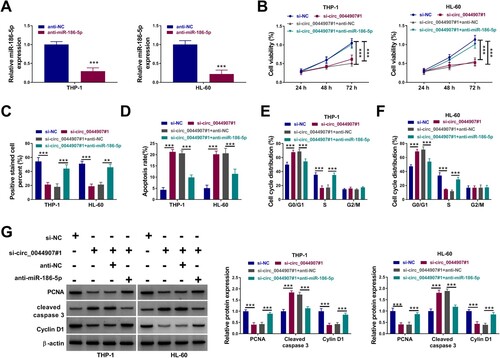
Figure 5. MiR-186-5p directly targeted KIT. (A) The putative binding sites of miR-186-5p in the 3’UTR of KIT. (B) Luciferase activity of KIT-3’UTR-WT/KIT-3’UTR-MUT reporter in AML cells co-transfected with miR-186-5p mimic or miR-NC. (C and D) Effects of miR-186-5p overexpression and knockdown on KIT protein levels in AML cells. (E) KIT protein levels in AML cells with si-NC, si-circ_0044907#1, si-circ_0044907#1 + anti-NC, or si-circ_0044907#1 + anti-miR-186-5p. (F) Relative expression changes of KIT mRNA in AML patient-derived BM. (G and H) Pearson’s correlation coefficient analysis of the correlation between KIT mRNA and miR-186-5p or circ_0044907 in AML patient-derived BM. (I and J) Detection of KIT protein levels in AML patient-derived BM and AML cells. *P < 0.05 and ***P < 0.001.
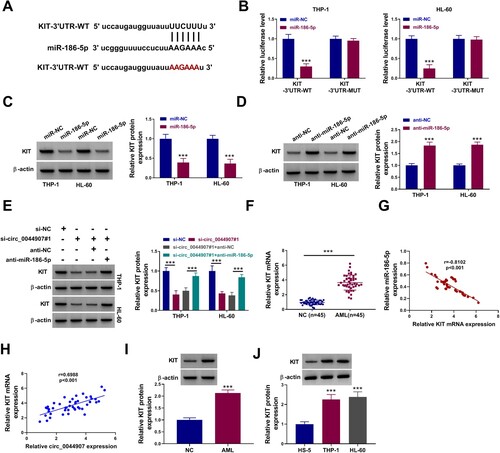
Figure 6. MiR-186-5p mediated AML cell proliferation and apoptosis via negatively regulating KIT expression. (A) Western blotting analysis of the transfection efficiency of the KIT overexpression plasmid. (B-F) Determination of cell viability (B), proliferation (C), apoptosis (D), and cycle progression (E and F) in AML cells with miR-NC, miR-186-5p, miR-186-5p + vector, or miR-186-5p + KIT. (G) Detection of PCNA, Cyclin D1, and Cleaved caspase 3 protein levels in the above AML cells. **P < 0.01 and ***P < 0.001.
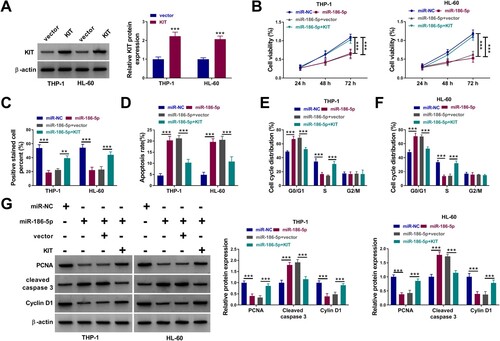
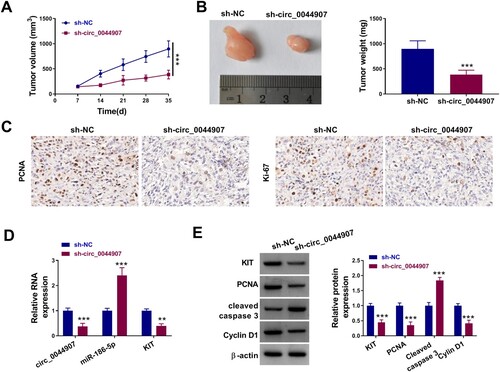
Figure 7. Circ_0044907 facilitated AML cell in vivo. (A) The tumor volume of sh-circ_0044907 and sh-NC groups. (B) Representative images of xenograft tumors in the above two groups (Left). The average weight of xenograft tumors in the above two groups (Right). (C) Representative images of IHC staining for PCNA and Ki-67 in xenograft tumors in the above two groups. (D) Expression levels of circ_0044907, miR-186-5p, and KIT mRNA in xenograft tumors in the above two groups. (E) Protein levels of Cleaved caspase-3, KIT, PCNA, and Cyclin D1 in xenograft tumors in the above two groups. **P < 0.01 and ***P < 0.001.