Figures & data
Figure 1. Diagram of the procedure for the 24 rare variant association studies (RVAS’s) between candidate variants and clinical severity in patients with Hb E/beta-thalassemia. Variants with passing the Variant Quality Recalibration Score (Pass) and/or Excess het filter (Excess het), different allele frequencies, and different predictions [high impact, moderate impact to protein by variant effect predictor (VEP), and moderate impact with deleterious effect by meta-logistic regression (meta-LR)] were grouped for analysis.
![Figure 1. Diagram of the procedure for the 24 rare variant association studies (RVAS’s) between candidate variants and clinical severity in patients with Hb E/beta-thalassemia. Variants with passing the Variant Quality Recalibration Score (Pass) and/or Excess het filter (Excess het), different allele frequencies, and different predictions [high impact, moderate impact to protein by variant effect predictor (VEP), and moderate impact with deleterious effect by meta-logistic regression (meta-LR)] were grouped for analysis.](/cms/asset/e2b651cd-9261-4b62-ac96-88c9fb1990e8/yhem_a_2187155_f0001_oc.jpg)
Table 1. Clinical and laboratory information of mild and severe patients were compared according to the scoring system.
Figure 2. Sanger sequencing revealed in cis interaction of IVS I-7 (A > T) (β+) and codon 26 (G > A) (βE) coinherited with in trans codon 26 (G > A) (βE).
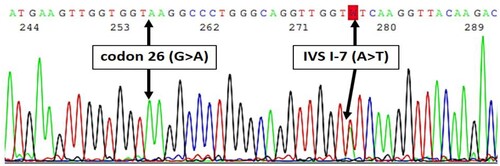
Figure 3. Diagram showing 53 out of 338 patients with phenotype of thalassemia but no thalassemia genotypes analyzed by whole exome sequencing (WES).
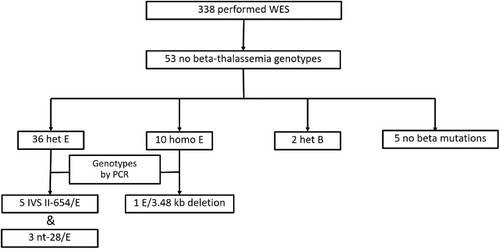
Figure 4. Whole exome sequencing showing the number of reads covered on IVS-II-654 and nt-28 mutations. The number of reads was too low. Therefore, the mutations could not be confidently called by whole exome sequencing. (A) The IVS-II-654 mutation was located in the second intron. (B) The nt-28 mutation was located in the promoter. Both mutations were confirmed by PCR.
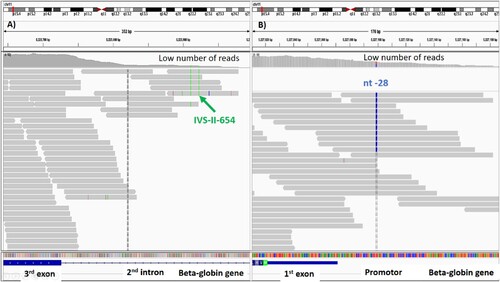
Figure 5. The sample was called as homozygous Hb E by whole exome sequencing, but it was compound heterozygous mutation of 3.48 kb deletion and codon 26 (G > A) (βE) by PCR. Codon 26 (G > A) (βE) from the remaining chromosome was called from all reads.

Table 2. Results of 24 rare variant association studies between variants within the top six genes and clinical severity of patients with beta-thalassemia.
Table 3. Number of cases of mild and severe phenotypes with variants in known modified genes of beta-thalassemia.
Table 4. Clinical and laboratory information of the six patients with heterozygous KLF1 variants.
Table 5. Predicted consequences and allele frequencies of the four KLF1 variants.
