Figures & data
Figure 1. Hydroxyproline content in leaves of sorghum as a reaction to anthracnose caused by C. sublineolum. The content of HRGPs, estimated as the content of hydroxyproline (Hyp) in (a) the resistant genotype SC146, (b) the intermediately resistant genotype SC326 and (c) the susceptible genotype BTx623 of sorghum in plants either treated with distilled water (controls) or inoculated with C. sublineolum. Statistical comparisons were made between the inoculated and control samples at each time point in each genotype. Data are the means of two replicates. The number of asterisks indicates the degree of significance. NS: non-significant difference, ***: significant at p≤0.001, **: significant at p≤0.01, *: significant at p≤0.05. Bars represent standard deviations.
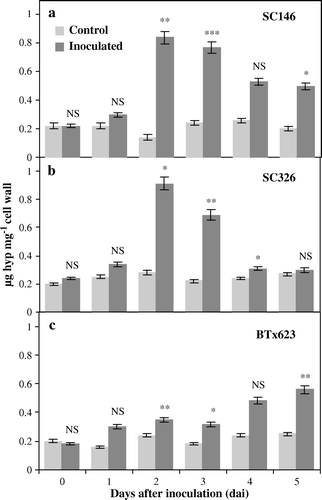
Figure 2. Western blot analysis showing the temporal accumulation of HRGPs in sorghum using the pAB-P/HRGPs antibody from pearl millet. (a) HRGP accumulation in sorghum leaves of the resistant genotype SC146, the intermediately resistant genotype SC326 and the susceptible BTx623 as a response to C. sublineolum infection. Lane 1: genotype SC146 control; Lane 2: genotype SC146 inoculated; Lane 3: genotype SC326 control; Lane 4: genotype SC326 inoculated; Lane 5: genotype BTx623 control and Lane 6: genotype BTx623 inoculated. (b) Quantification of intensities of the ~45 kDa, 17 kDa and 14 kDa bands using the Bioprofile Image Analysis System. Bars represent standard deviations.
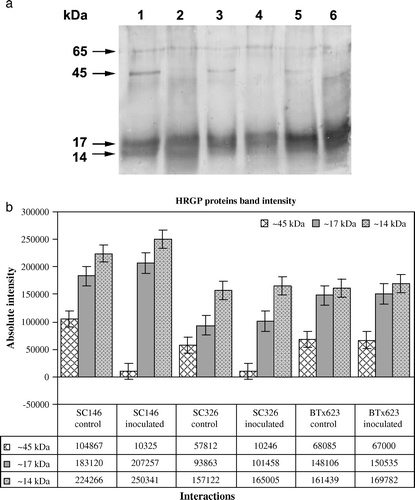
Figure 3. Light micrographs of H2O2 accumulation at infection sites of C. sublineolum in leaves of sorghum. H2O2 accumulation is seen by DAB-staining at infection sites in (a) the resistant genotype SC146, whereas in (b) no staining is seen in the susceptible genotype BTx623 where successful development of pathogen took place. Scale bars = 50 µm.
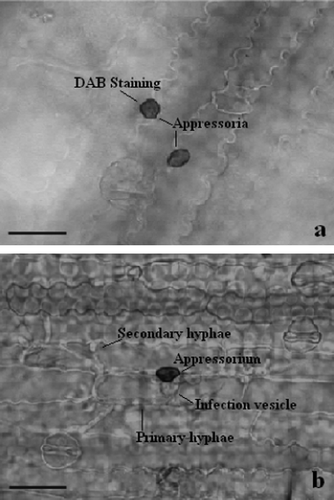
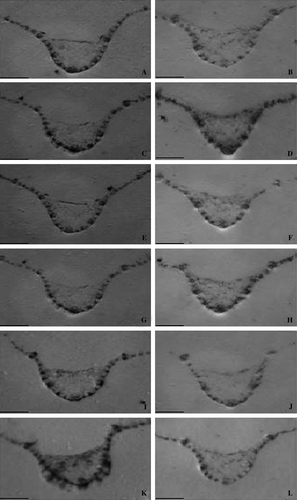
Figure 4. Tissue print immunoblot localization of HRGPs in sorghum leaves either inoculated with C. sublineolum or left untreated (controls), using pAB-P/HRGPs polyclonal antibodies to localize HRGPs. Tissue prints were made from cross sections of leaves from the resistant genotype SC146 (B: uninoculated, D: inoculated), the intermediately resistant genotype SC326 (F: uninoculated, H: inoculated) and the susceptible genotype BTx623 (J: uninoculated, L: inoculated). The total basal level of protein in the resistant genotype SC146 (A: uninoculated, C: inoculated), the intermediately resistant genotype SC326 (E: uninoculated, G: inoculated) and the susceptible genotype BTx623 (I: uninoculated; K: inoculated) was stained with Coomassie brilliant blue. Scale bars = 3 mm.