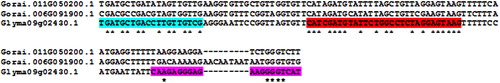
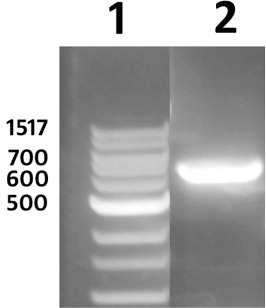
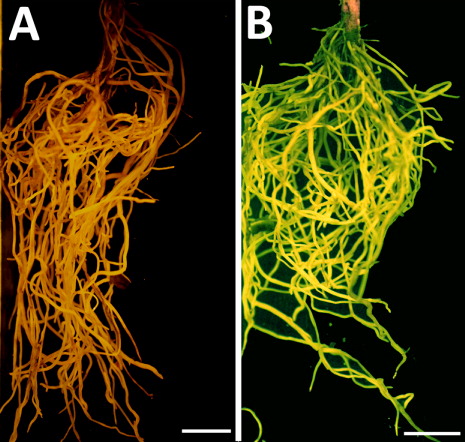
Figures & data
Table 1. PCR primers. The primers used are A. rhizogenes VirG (Ar-VirG), G. hirsutum S21 (Gh-S21), G. max NPR1-2 (Gm-NPR1-2), and eGFP.
![Figure 1. pRAP15 vector. Legend: open reading frame (ORF), pink; ORF1, homologous to partitioning protein A (ParA); ORF2, no significantly homologous leader sequence 5′ fused to the Reporter gene (enhanced green fluorescent protein [eGFP]), green. ORF3, homologous to resolvase; ORF4, homologous to replicase A (RepA); ORF5, homologous to tetracycline resistance gene (TetR); ORF6, homologous to aminoglycoside adenyltransferase. Origin of replication (pBR322), black. Other gene (ccdB, attR1, attR2), magenta. Selectable marker (chloramphenicol [CAT]), orange. Terminator (cauliflower mosaic virus [CaMV] 35S), brown. Blue lettering, unique restriction sites.](/cms/asset/2ed2dfa6-cdc6-45a6-8e45-c20e46dec7f4/tjpi_a_1005181_f0001_c.jpg)
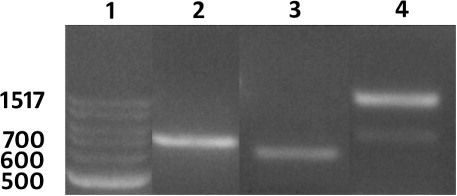

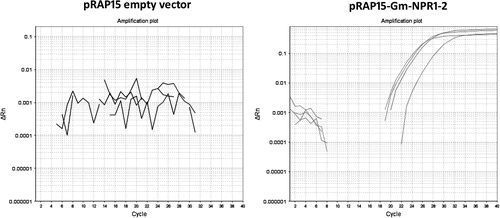
Table 2. A time line for production of composite G. max and G. hirsutum plants with transformed roots.