Figures & data
Table 1. Primers used in present study.
Table 2. The overall information on HHP proteins from different crops.
Figure 1. Amino acid sequence alignments of heptahelical proteins (HHPs) from Pak-choi and other crops. BcHHP3 was isolated in the study and some other crop proteins were selected from GenBank (A). Less conserved residues, highly conserved residues and perfectly matched residues are represented by a green box, a pink box and a black box, respectively (B).
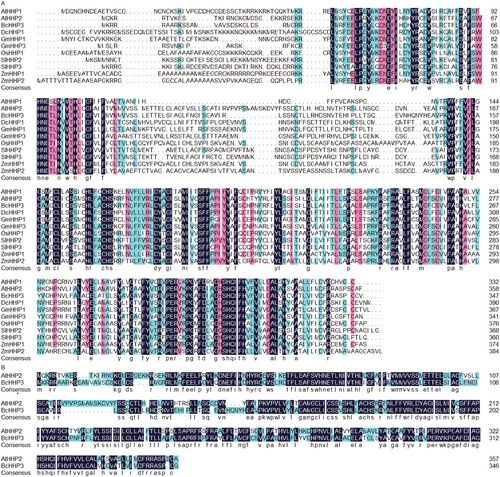
Figure 2. Phylogenetic and structural analyses of the putative BcHHP3 protein and other HHPs available from GenBank and other databases were carried out. The unrooted tree on the left of the figure is based on an alignment of the full-length protein sequences and was constructed by the maximum likelihood method. The schematic structures of HHP proteins in the selected plants are shown on the right side of the figure (the left panel). The schematic structure of some of the HHP proteins in the selected plants is shown. Different types of patterns are represented by boxes of different color types. The same box type in different proteins indicates the same motif (the right panel). Species are as follows: At, Arabidopsis thaliana; Bc, Brassica campestris; Dc, Daucus carota; Gm, G. max; Os, O. sativa; Sl, Solanum lycopersicum; Zm, Z. mays.
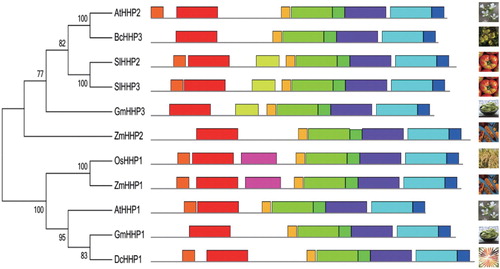
Figure 3. Comparison of the secondary structure of the BcHHP3 protein. Helix, turn, strand and coil were indicated in line with decreased length in turn, respectively (A). Helix, turn, strand and coil were indicated with blue, red, green and purple lines, respectively (B).
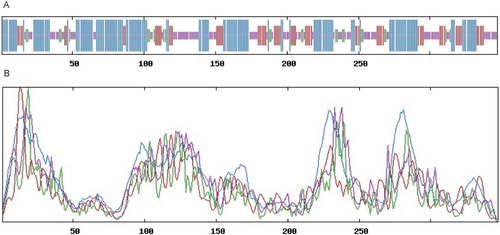
Figure 4. The predicted 3D structure of the BcHHP3 protein and the related information. The predicted 3D structure of the BcHHP3 protein (A). Local Quality Estimate of the BcHHP3 protein (B). Global Quality Estimate of the BcHHP3 protein (C). Comparison with Non-redundant Set of PDB Structures (D).
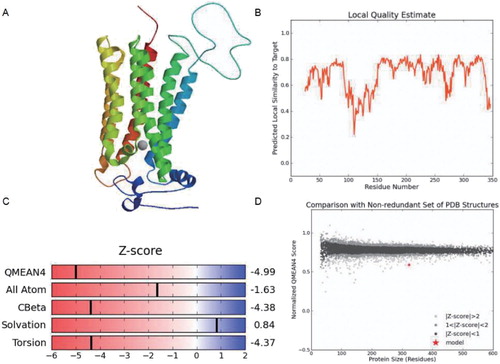
Figure 5. qRT-PCR analysis of expression patterns on BcHHP3 in Pak-choi under abiotic stress. BcHHP3 expression under cold treatment (A). BcHHP3 expression under salt treatment (B). BcHHP3 expression under ABA treatment (C). BcHHP3 expression under SA treatment (D). The transcript levels of BcHHP3 relative to those of BcACTIN were quantified using 2−ΔΔCT ; ΔΔCT = ΔCT (treated sample) − ΔCT (untreated sample), ΔCT = CTtarget − CTBcACTIN. Each column represents the average and standard deviation of three replicates. *.01 < P < .05, **P < .01, significant differences between treatments, respectively (ANOVA calculated using SPSS). Each bar value represents the Means ± SEs of the three replicates.
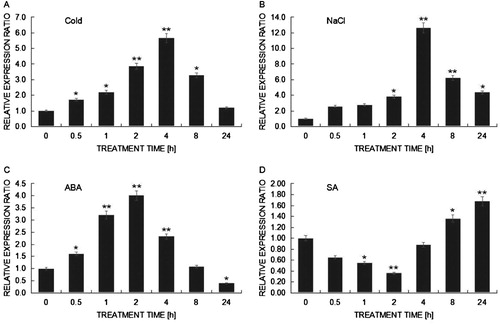
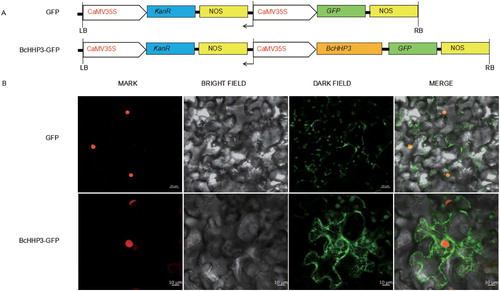
Figure 6. Subcellular localization of BcHHP3 in tobacco cells. Constructs were used in the experiments (A). BcHHP3-GFP were transferred to tobacco cells through injecting leaves to test and verify the subcellular localization of BcHHP3 (B). The upper panel shows mark, bright field, dark field and merge images of the GFP control; the lower panel shows mark, bright field, dark field and merge images of BcHHP3-GFP.