Figures & data
Figure 1. Phenotypes of 35S:ER/35S:OFP15 and 35S:ER/35S:OFP16 transgenic plants. Lower, morphology of the Col wild type, er mutant, 35S:OFP15, 35S:OFP16, 35S:ER/35S:OFP15 and 35S:ER/35S:OFP16 transgenic plants. Upper, close view of the silique end of the fourth silique from main inflorescence of Col wild type, er mutant, 35S:OFP15, 35S:OFP16, 35S:ER/35S:OFP15 and 35S:ER/35S:OFP16 transgenic plants. All the plants were grown side by side in soil pots. Plants ∼7-week-old were photographed by using a digital camera.

Figure 2. Phenotypes of er ofp15 and er ofp16 double mutants. (A) Morphology of the Col wild type, er, ofp15 and ofp16 single mutants, and er ofp15 and er ofp16 double mutants. All the plants were grown side by side in soil pots. Plants ∼4-week-old were photographed by using a digital camera. (B) The fourth silique from main inflorescence of Col wild type, er, ofp15 and ofp16 single mutants, and er ofp15 and er ofp16 double mutants. All the plants were grown side by side in soil pots. Morphology of siliques in ∼7-week-old plants were observed and photographed by using a digital camera.

Figure 3. Expression of OFP15, OFP16 and OFP18 in er mutant seedlings. Total RNA was extracted from seedlings of the Col wild type and er single mutant, and RT-PCR was used to examine the expression levels of OFP15, OFP16 and OFP18. The expression level of ACT2 was used as a control.
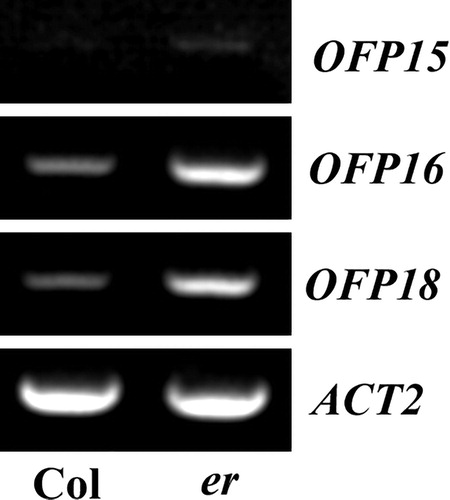
Figure 4. Effects of transcriptional activator VP on the functions of Class III OFPs. (A) Subcellular localization of Class III OFP proteins. Plasmid DNA of corresponding OFP effector was transfected into protoplasts, the transfected protoplasts were incubated at room temperature under darkness for 20–22 h, and then GFP fluorescence was examined and photographed under an Olympus FV1000 confocal microscope. (B) Transcriptional activities of VP fused Class III OFP transcription factors. Plasmid DNA of the Gal4:GUS reporter and corresponding OFP effector was co-transfected into protoplasts, the transfected protoplasts were incubated at room temperature under darkness for 20–22 h, and then GUS activity was measured by using a SynergyTM HT fluorescence microplate reader (BioTEK). Data represent the mean ± SD of three repeats. The experiment was repeated three times with similar results. (C) Phenotypes of 35S:OFP16VP and 35S:OFP18VP transgenic plants. All the plants were grown side by side in soil pots. Plants ∼3-week-old were photographed by using a digital camera.
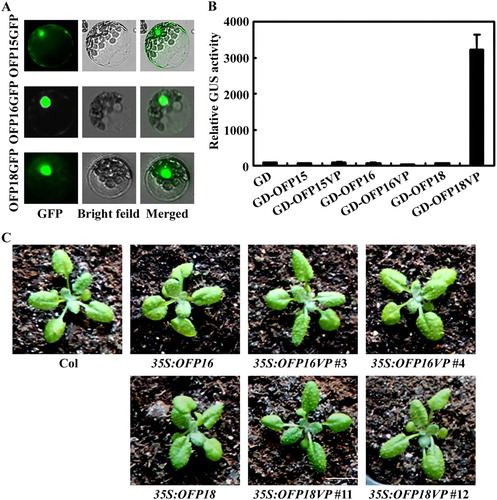
Figure 5. Effects of amino acid substitution of the putative MAPK phosphorylation sites on the transcriptional activity of OFP15 and OFP18. (A) Putative MAPK phosphorylation sites in OFP15 and OFP18. Stars indicate the putative phosphorylation sites. (B) Transcriptional activities of putative phosphorylation sites substituted OFP15 and OFP18. Plasmid DNA of the LexA-Gal4:GUS reporter, transcription activator LD-VP and corresponding OFP effector was co-transfected into protoplasts, the transfected protoplasts were incubated at room temperature under darkness for 20–22 h, and then GUS activity was measured by using a SynergyTM HT fluorescence microplate reader (BioTEK). Data represent the mean ± SD of three repeats. The experiment was repeated three times with similar results.
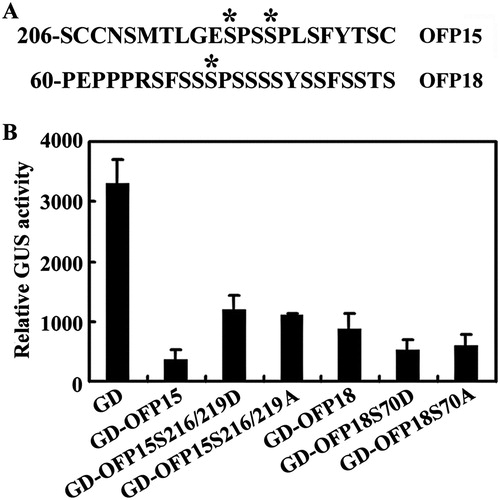
Figure 6. Phenotypes of transgenic plants overexpressing phosphorylation sites substituted OFP15. (A) Morphology of the Col wild type, 35S:OFP15, 35S:OFP15S216/219D and 35S:OFP15S216/219A transgenic plants. All the plants were grown side by side in soil pots. Plants ∼4-week-old were photographed by using a digital camera. (B) The fourth silique from the main inflorescence of the Col wild type, 35S:OFP15, 35S:OFP15S216/219D and 35S:OFP15S216/219A transgenic plants. All the plants were grown side by side in soil pots, the morphology of siliques in ∼7-week-old plants was observed and photographed by using a digital camera.
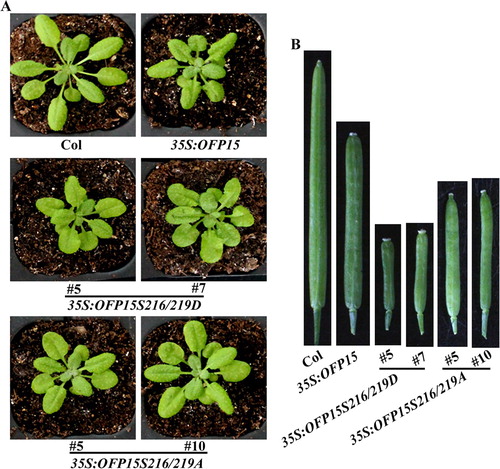
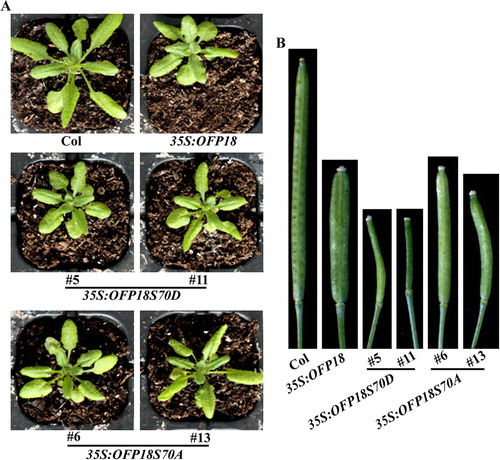
Figure 7. Phenotypes of transgenic plants overexpressing phosphorylation sites substituted OFP18. (A) Morphology of the Col wild type, 35S:OFP18, 35S:OFP18S70D and 35S:OFP18S70A transgenic plants. All the plants were grown side by side in soil pots. Plants ∼4-week-old were photographed by using a digital camera. (B) The fourth silique from the main inflorescence of the Col wild type, 35S:OFP18, 35S:OFP18S70D and 35S:OFP18S70A transgenic plants. All the plants were grown side by side in soil pots, the morphology of siliques in ∼7-week-old plants was observed and photographed by using a digital camera.