Figures & data
Table 1. Basic information list of the MATE proteins in pigeonpea genome Abbreviations: pI, isoelectric point; Mw, molecular weight; aa, amino acid.
Figure 1. The phylogenetic tree of MATE proteins from C. cajan and other plant species. The phylogenetic tree was constructed with the MEGA 6.0 program by the neighbor-joining method. The Bootstrap value (percentage) (1000 repetitions) is displayed on the node. Different subgroups are highlighted using different colors.
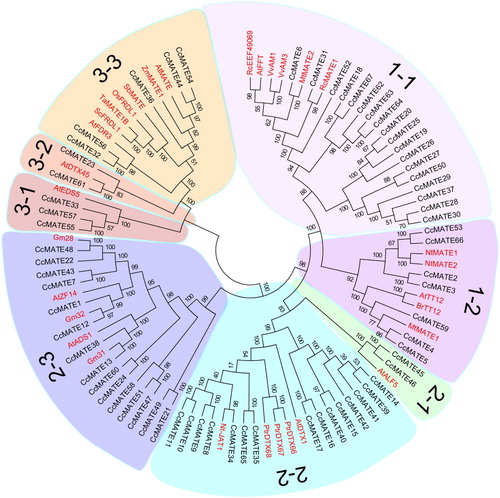
Figure 2. Predicted structures of MATE proteins. The structures of 9 MATE proteins were predicted to a >90% confidence interval.

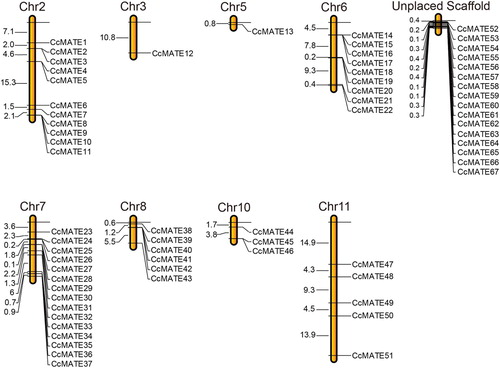
Figure 3. The distribution of CcMATE genes in pigeonpea chromosomes. The number of MATE genes in each chromosome are shown in the top. The CcMATEs were named CcMATE1 to CcMATE67 based on their code number in the pigeonpea genome.
Figuer 4. Conserved domains of the MATE proteins in pigeonpea. A. The motif details of five highly conserved domains identified in CcMATE proteins and the logos of these domains created using the MEME program. B. The CcMATE proteins are listed according to the order of subfamily 1–3 in the phylogenetic tree. Each motif is represented by a colored box. He length of the motif can be estimated using the bottom scale (unit: amino acids).
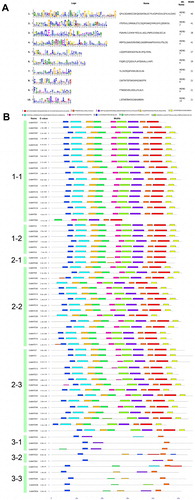
Figure 5. Expression profiles of 8 CcMATE genes in four pigeonpea tissues. The color bar represents the expression value. Each group has three biological replicates.

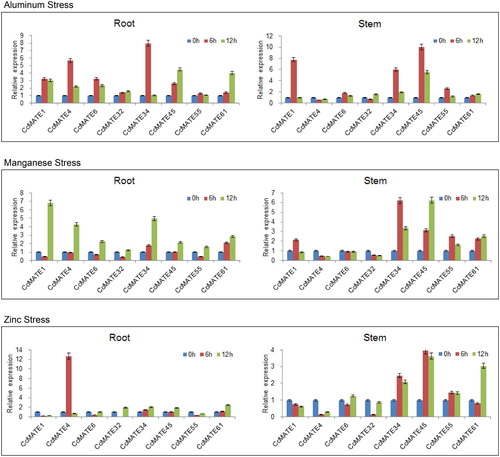
Figure 6. Relative expression levels of 8 CcMATE genes in response to three metal stress, as determined by quantitative real-time PCR. Eight representative CcMATE genes were selected from the eight subgroups. The expression patterns of these CcMATEs in pigeonpea roots and stems were examined under aluminum, manganese and zinc stress. The roots and stems were harvested for 6 and 12 h after treatment. Each group has three biological replicates.